
Precision medicine is a gradually developing field that utilizes specific genetic information of patients to devise treatment methodology. Cancer is no exception, in a majority of the cases, there are genetic mutations involved in the incidence and maintenance of cancer in addition to external/environmental factors. Understanding the underlying genetic aberration(/s) in individual cancer patients can fine-tune their treatment strategies- can bring us one step closer to determining the right therapy for individual patients. Precision medicine is confined because of lack of enough knowledge of the molecular/genetic bases of different types of cancer; even individuals with the same kind of carcinoma may contain distinct, dissimilar gene mutations and dissimilar biomarkers. Current methods of treatments are more like ‘one size fits all’ and constitute primarily of chemotherapy with certain combinatorial drugs, with some minimal targeted treatment options (precision therapy). Precision medicine is picking up speed at present; however, the standard technique for determining the gene alteration/molecular basis of picking up speed at present; however, the standard technique for determining the gene alteration/molecular basis of cancer is tissue biopsy.
Tissue biopsy is the core of determining underlying adverse genetic mutation for cancer incidence and progression, tumor growth monitoring, therapy response, and relapse. It has its shortcomings though- it is invasive, and the analysis takes a long time (several weeks). Since it is an invasive technique, it is somewhat tricky to perform follow-up analyses, cannot be pursued if the tumor is inaccessible, is potentially complicated, and cannot be utilized if cancer has worsened. The common types of tissue biopsy- encompassing surgical biopsy, endoscopic biopsy, bone marrow biopsy, needle biopsy, and skin biopsy- are being relied upon massively for monitoring cancer and devising the correct course of directed therapy. These traditional biopsy techniques also entail the risk of limited information (localized sampling) regarding tumor heterogeneity- cancer cells are a heterogeneous population, and this dynamicity evolves during every stage of tumor progression towards metastasis. In addition to all these
factors, there is an inherent risk of preserved tissue samples- a process that involves chemicals that may result in alteration of biomolecules in the tissue, thereby, adversely affecting actual results.
To overcome these hindrances, a plausible technique that can help in continuous analysis of cancer progression and treatment response, without being affected by the inaccessibility of the tumor, is a liquid biopsy [1]. A method of isolation/extraction of tumor-shed biomolecules, cells, and exosomes from body fluids, and their subsequent characterization to determine the transcriptomic and proteomic profile is known as liquid biopsy. Liquid biopsy is gaining popularity at present as a crucial tool that not only bypasses the problems mentioned above in cancer therapy; it is non invasive too- one of the most significant advantages. Molecular genomic analyses and immunotherapy can be paired with a liquid biopsy from patients within a much shorter time in comparison to traditional tissue biopsy, as evident from current literature. Different types of body fluids can be utilized as sources of the biopsies- blood, urine, cerebrospinal fluid (CSF) and even saliva; blood is most commonly used among all.
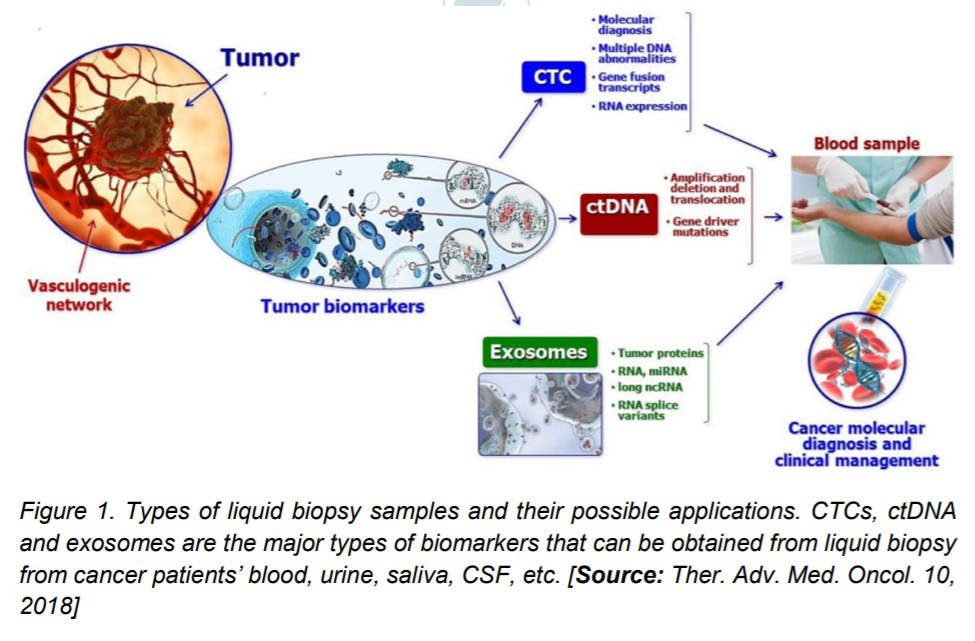
The components that are characterized by blood sample biopsy are circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and exosomes. These biomarkers are analyzed using myriad techniques to either determine tumorspecific DNA mutations, or cell surface receptors on tumor cells or for understanding the epigenetic cues of cancer progression. Initial investigations were more restricted to determining the number of CTCs that radically increases with tumorigenesis. This technique has further given way to characterization of the biomolecules on and within the cellsDNA and specific proteins/ protein receptors. The continual shedding of cells, DNA, and vesicles by primary and metastatic tumors makes it convenient for the seamless isolation and investigation of these biomarkers. Various techniques have been used to isolate the CTCs from the bloodstream of cancer patients.
Circulating tumor cells (CTCs)
First reported in 1869, these cells generate secondary tumors and therefore, cause metastasis from primary tumor because of their body fluidmediated mobility. The tumors shed the CTCs because of high turnover of cancer cells. The CTCs possess highly diversified surface proteins/receptors that lead to their heterogeneity at and across different stages of cancer, they are also rare in the bloodstream of patients- requires highly sensitive and specific detection and isolation techniques [1]. Over the years, various research groups have worked towards refining the detection methods of the heterogenous CTC population in tumorafflicted patient blood samples. Currently, the standard techniques are based on the biological properties of these cells- surface antigens/receptors, or their physical features, including density, size, and inertia. The biological sorting and detection depend on using specific antibodies against specific cancer-type dependent surface proteins, and sometimes, the immunoaffinity is combined with magnetic separation of the antibody bound CTCs [2, 3]. CellSearch, the only FDA-approved CTC detection kit utilizes this immunomagnetic technique and has successfully isolated CTCs (epithelial origin) that are EpCAM+, CD45- (leukocyte marker), CK19+, besides other cell surface antigens. This kit has been successfully used in isolating CTCs in clinical trials for breast cancer, colorectal cancer, and prostate carcinoma. Immunostaining of the CTCs follows their immunomagnetic detection and helps in differentiating the blood cells from the CTCs, thereby enumerating them amongst the leukocytes. Besides the immunomagnetic method, high throughput imaging, leukocyte depletion, density/size (the CTCs are bigger compared to blood cells), and adhesionbased isolation methods are used to detect CTCs. More sophisticated technologies are developing that not only enumerate the CTCs, but also characterize the biomolecules inside them- DNA, RNA, vesicles, and proteins.
Circulating tumor nucleic acids (ctNAs)
Constituting of tumor-derived DNA and RNA, these biomarkers are an essential category of cancer prognosis factors [2, 4, 5]. The tumor cfDNA fragments have an average length of about 150 bp, constitute of only about 0.1% to 10% of the total cfDNA in circulation, and have a shorter lifespan in comparison to normal apoptotic/necrotic cell-shed cDNA. These DNA molecules are believed to have metastatizing potential, as reported in several studies, including colorectal cancer and colon cancer. Like CTCs, an increase in the number/amount of these tumor-shed cfDNA signifies poor prognosis and thereby poor overall survival. Most of the pre-clinical and clinical studies currently use blood as a source of ctDNA for extraction and analysis. Isolation of the tumor cfDNA is based on the following principlesmagnetic separation, electrical force, column-based affinity, fluorescent probes, or filtration [6]. Of late, nanotechnological advancements have aided in isolation of pure ctDNA from the blood plasma of breast and lung cancer patients. Scientists and clinicians use either these nano-tech methods or available commercial kits and can investigate the whole genome analyses by omic approaches; they also study specific genes for their expression levels, mutations and epigenetic signatures like methylation pattern. The various methodologies include RNA-Seq, NGS technologies, digital PCR (dPCR), peptide-nucleic acid-locked clamp PCR (PNA-LNA), allele-specific PCR, and others; all of these techniques have their advantages and limitations, however, combining multiple methodologies provides a more comprehensive picture of the tumor microenvironment [7]. Sorensen et al. used the tumor cfDNA isolated from plasma to investigate specific drug-resistant mutations in the EGFR gene by allele-specific PCR, Oxnard et al. utilized digital PCR to isolate the exon 19 deletion patients of the advanced NSCLC, and Sozzi compared the cfDNA levels in NSCLC patients with healthy individuals using the plasma-derived cfDNA fragments. So, similar to the CTCs, the ctDNA is an exhaustive source of information that is pushing cancer therapy, disease monitoring, remission, and/recurrence to new heights. In spite of providing a wealth of knowledge to help the clinicians and medical professionals, tumor cfDNA extraction has an inherent variation in extraction amount among different samples; the investigation on finding the known mutations in various cancers cast a shadow on the versatility of the cfDNA application too- it lacks knowledge about possible new mutations associated with the cancer because of narrow target region. Therefore, further refining of techniques is underway to improve uniformity of extraction and a broader gene interrogation region has been
added to focus on a genome-wide scale to generate a vast repertoire of the associated mutations (digital karyotyping and personalized analysis of rearranged ends/PARE). Newman et al. investigated plasma obtained tumor cfDNA for sets of mutations on a genome-wide scalevariations that are common in NSCLC. Though comprised of small sample size, there was a perfect correlation of cfDNA result with stage II, III, and IV patients.
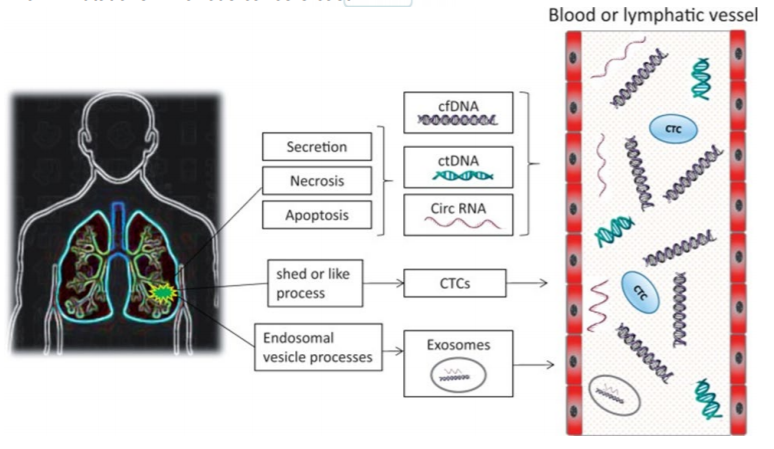
Figure 2. Sources of ctDNA. Healthy cells, necrotic or apoptotic cells can shed cfDNA. The cfDNA can also secreted passively into the bodyfluids. However, tumor cells shed cfDNA (called ctDNA) in circulation at a much higher rate than normal, as a result of incomplete processing and rapid tumor cell turnover. Exosomes also serve as a source of ctDNA in cancer patients. [Source: Experimental Biology and Medicine, 243(3):262-
271, 2018]
As biomarkers, microRNAs(miRNAs) and long non-coding RNAs (lncRNAs) are pivotal to cancer progression, since tumor growth and spread require upregulation of oncogenes and gene expression machinery, while simultaneously downregulating tumorsuppressing genes such as p53 [2]. In addition to the circulated RNA in the body fluids, exosomes can carry the RNAs to a destination- a secondary or tertiary site of the tumor. The lncRNAs are crucial for regulating gene expression: mRNA splicing and stability. miRNAs regulate the expression of about 30% of human genes (protein-coding genes) and there are reports of non-exosomal miRNA in the bloodstream that become stabilized by binding RNA-binding proteins or highdensity lipoproteins. Therefore, these non-coding RNAs are a promising biomarker for understanding tumor growth as well as treatment outcome.
Exosomes
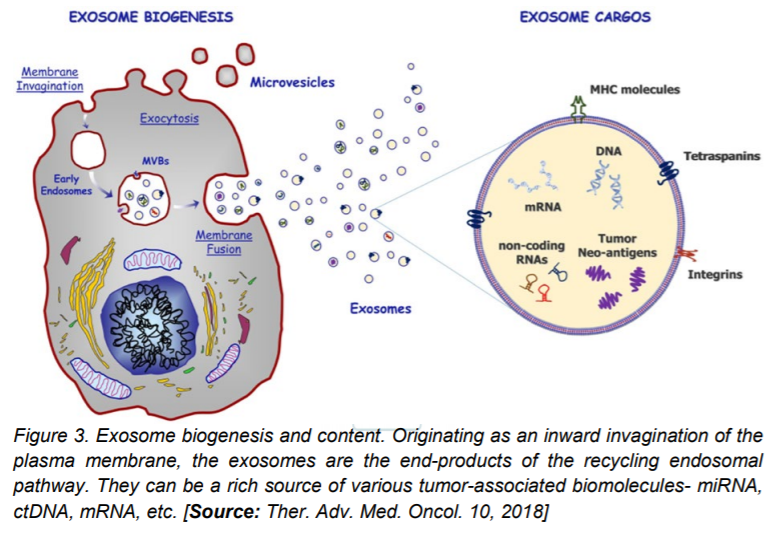
A vital mode for intercellular communication between cells, the exosomes carry biomolecules including membrane and non-membrane proteins, microRNAs, long non-coding RNAs, single or double-stranded DNAs, and lipids [1, 6]. These players are emerging as key drivers in chemo-resistance and they mediate tumor angiogenesis, proliferation, and metastasis. Abundantly found in patient body fluids like plasma, saliva, ascites, and urine, the exosomes are known to contain a conserved set of the membrane and cell surface proteins, namely CD9, CD81, and CD63, though studies from different groups show cell type or tissue-type specific protein content of exosomes. Traditional methods of extraction involving size dependent filtration has given way to a more sophisticated methodology- the centrifugal microfluidic platform (microfluidic centrifugal nanoparticles separation and isolation or µCENSE) that reduces the total time of exosome extraction considerably. Immunocapture, with antibodies against CD63 or CD4 are also in use to extract exosomes from human sera that can be successfully utilized for downstream analyses.
Further technological upgrades have now added the use of electrical field in exosome isolation. Simultaneous detection, enrichment, and investigations are now possible by commercially available ‘on-chip’ that combines immunocapture and electrochemical signaling. Various investigators, including Doldan et al., Wang et al., and Ko et al. have refined this advanced technique further by enhancing the sensitivity of the detection system by several folds. For analyses of the contained biomolecules, the isolated exosomes are tested for the presence of marker proteins; genome-wide analyses are also performed to obtain a bigger picture of the tumor spread and/therapy response in patients.
It is thrilling to be able to study cancer- a debilitating and fatal disease- at the ease of a blood draw or collecting a urine sample from the patient [8, 9]. This remarkable advancement is a step closer to devising patient need-specific treatment strategies, something highly relevant in cancer therapy. Constant evolution of cancer markers throughout its growth and presence or absence of similar markers in different patients with the same cancer type, are two of the biggest hindrances on the path of recovery from cancer. Affordability, accessibility, shorter turnover time of detection, and analyses are all contributory to the central idea of personalized precision medicine. Though the liquid biopsy technologies prevalent at present are substantially informative, they still have to be combined with established tumor detection and analytical procedures that include imaging and tissue biopsy; the sensitivity and specificity can also be concerns, depending on the biomarker in question. Therefore, it is imperative that clinicians and pre-clinical researchers invest their resources extensively to improve this hallmark tumor monitoring method- liquid biopsy- to develop it as a comprehensive tool for cancer detection and monitoring technology soon.
Author’s Biography
Dr. Progga Sen is a postdoctoral research fellow at the Stanford University School of Medicine, working at the Veterans Affairs Health Care System Palo Alto. She focuses on understanding the mechanism of substrate-mediated regulation of long-chain fatty acyl-CoA synthetase family member 4 (ACSL4), her research has implications in liver disorders including non-alcoholic fatty liver disease (NAFLD) and non-alcoholic hepatosteatosis (NASH). Besides research, she has a strong inclination towards writing engaging blogs and articles. Recently, she published her blog on the award-winning blog website SCOPE (published by Stanford School of Medicine). Progga completed her Ph.D. at the Wayne State University from the Department of Biology and has presented her research at several annual symposia and research retreats during her Ph.D. Her keen interest in science communication and education has been reinforced by her appointment as a Graduate Teaching Assistant. She intends to work towards improving human health and quality of life. To that end, she is leveraging her extensive academic background and strong communication skills to successfully convey scientific discoveries and advancement tailored for intended target audience. She can be reached at progga.sen@gmail.com
References
1. Raffaele Palmirotta, et al. (2018). Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol., 10: 1758835918794630.
2. Jose Marrugo-Ramirez, Mir M, and Samitier J. (2018). Blood-based cancer biomarkers in liquid biopsy: A promising non-invasive alternative to tissue biopsy. Int. J. Mol. Sci. 19 (10).
3. Sanne de Wit et al. (2019). Single tube liquid biopsy for advanced non-small cell lung cancer. Int. J. Cancer, 144(12):3127-3137.
4. Donald J. Johann Jr, et al. (2018). Liquid biopsy and its role in an advanced clinical trial for lung cancer. Experimental Biology and Medicine, 243(3):262-271.
5. Jacqueline Howard (2019). Using blood, saliva, urine to detect cancer: Scientists’ ‘Holy Grail’. www.cnn.com.
6. Ramanathan Vaidyanathan et al. (2018). Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab on a Chip, 19(1):11-34.
7. Junaid Ansari, et al. (2017). The liquid biopsy in lung cancer. Genes & Cancer, 7 (11-12).
8. Alissa Poh. (2019). Liquid biopsy holds its own in NSCLC. Cancer Discovery.
9. Della O’Hare (2018). A blood test may one day detect early stages of cancer. Discover Magazine, June Issue.