Chromothripsis: A Cause of Genome Instability in Cancer
Ishita Agrawal*
Molecular and Human Genetics, Banaras Hindu University, Varanasi – 221005, UP, India
DOI: https://doi.org/10.37756/bk.21.3.4.1
ABSTRACT
Cancer is a term given to uncontrolled cell growth, which is the result of accumulation of genetic changes during cell division. It can occur due to both genetic and environmental reasons. One such genetic cause of cancer discovered recently is known as Chromothripsis. It is defined as the fragmentation and rearrangement of chromosomes. Researchers are still trying to find the true mechanism underlying Chromothripsis. Two models have been significantly described; first one is the ‘micronuclei hypothesis’, which occurs as a result of chromosome mis-segregation during Mitosis. Second model is based on the ‘telomere crisis’, which occurs due to faulty telomerase enzyme and cell cycle checkpoint pathway. In this letter, we have tried to give brief information about the mechanisms behind Chromothripsis and their role in causing genome instability leading to cancer.
Keywords: Cancer; Genome Sequencing; Chromothripsis; DNA Replication; Chromosomes; Telomere
Received: March 10, 2021
Revised Manuscript: March 20, 2021
Accepted: March 22, 2021
*E-mail: ibeau.in1@gmail.com
1. Introduction: Chromosome Shattering & Mechanisms in Chromothripsis
Years of research have shown that cancer is a genetic disorder. However, unlike germline mutations, these disorders are generally caused by somatic mutations in genes such as proto-oncogenes, tumor suppressor genes and DNA repair genes as well. Studies have shown that both environmental factors such as radiation, chemical mutagens and endogenous stressors such as water, reactive oxygen species can interfere with DNA replication, repair, and division pathways and lead to genomic instability which can cause Cancer. Lately, a novel form of genomic instability known as Chromothripsis, discovered in 2011 has been considered as a cause of cancer. Its literal meaning is ‘Chromosome shattering’. In the phenomenon involving Chromothripsis, a single catastrophic event leads to breaking of chromosome into numerous short pieces which then rearrange and join randomly by DNA repair processes such as non- homologous end joining [1-7].
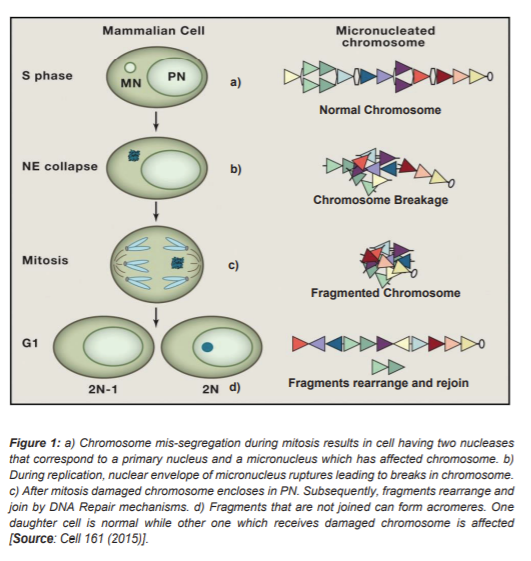
Studies to understand the cause of Chromothripsis have proposed two major mechanisms. Most extensively studied model is the ‘Micronuclei hypotheses’. Micronuclei are formed due to errors in chromosome segregation [8]. During anaphase stage of mitosis, paired chromosomes aligned at metaphase plate are separated which are then passed onto daughter cells. But, sometimes a chromosome or chromosome fragment lags or fails to align at chromosome plate and remains unattached to the spindle fibers. At the end of mitosis, if this chromosome is at sufficient distance from the metaphase plate it forms its own nuclear envelope (Figure 1) [9]. This small nucleus like structure formed during interphase is known as Micronucleus. This micronucleus is believed to be a major site of chromosome shattering and rearrangement known as ‘Chromothripsis’ [9].
Later studies showed that, when cells exit G1 phase nuclear envelope of micronucleus collapses and the fragmented or damaged chromosome gets reincorporated into Primary Nucleus [9]. The collapse of nuclear envelope of micronuclei, leads to lack of nuclear pore because of which important nuclear proteins are absent and replication of chromosomes present is either absent or delayed. On joining with Primary nucleus, these defects cause rearrangement of fragmented chromosomes. This rearrangement then leads to genomic instability and DNA damage [10].
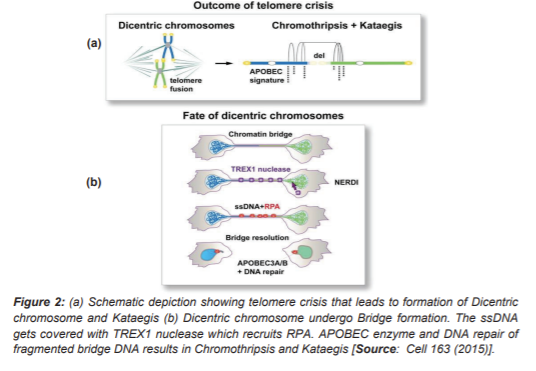
Another proposed model is ‘Telomere Crisis’, Figure 2 [11]. Telomere are tandem repeat sequences present at the ends of chromosomes and are responsible for protecting the ends of chromosomes and providing genomic stability. Each consecutive cell cycle leads to shortening of chromosomes which is a result of a phenomenon known as end replication problem. An enzyme ‘telomerase reverse transcriptase (TERT)’ solves this ‘end replication problem’. This enzyme activity is seen in germline cells whereas most somatic cells lack its expression. Loss of end repeat sequences or telomerase enzyme in single chromosome leads to formation of dicentric chromosomes which then undergo Breakage – Fusion Bridge cycles. These damages are generally not detected as they are identified by Cell cycle checkpoints and spindle check points (SAC) [11].
In tumorigenesis however, due to loss of p53 (a tumor suppressor gene) this telomere shortening leads to telomere dysfunction. In such cases, damaged chromosome bypasses cell cycle check points and telomerase lose the ability to bypass DNA repair mechanisms; the shortened chromosomes are joined via non- homologous end joining mechanism to form Dicentric Chromosomes and form Breakage –fusion bridge (BFB). Researchers showed that cells were made to express TERT; p53and Rb pathways were inhibited and telomere crisis was induced. This resulted in telomere dysfunction, activation of ATM repair pathway and disabled cell proliferation. Further, on attenuating telomere protecting gene TRF2, telomere fusion was observed which formed dicentric chromosome as expected [10, 11].
Researchers have also studied interplay of telomere crisis and Kataegis. Kataegis is a process of hypermutation caused by clustering of base substitutions. It does not generally occur in germline cells but in somatic cells. It has been showed that chromatin bridges recruit Replication Protein A (RPA) on extended ssDNA. These ssDNA are considered to be the target region for APOBEC enzyme, which converts cytosine to thymidine or uracil. This leads to the accumulation of substituted bases, which is a process known as Kataegis [11].
2. Chromothripsis in Cancer
Normally, damaged DNA is eliminated via apoptosis but in case of Chromotheripsis, cells survive based on affected genes present in that chromosome region. If the affected genes are important for pathways such as cell cycle regulation, DNA damage, repair, cell proliferation and apoptosis then, they will inhibit cell death and cell will survive [12].
2.1 Understanding the Mechanisms that Cause Cancer
The fragmented chromosomes can form acromeres (small, circular chromosome which lack centromere and telomere). Incorporation of above mentioned genes in acromeres can cause loss of tumor suppressor genes or can form Oncogenic fusions [12]. This is observed in cases of oesophageal adenocarcinoma and acute myeloid leukemia. In acute myeloid leukemia, this is most prevalent in chromosome 17 [12].
Another mechanism involves DNA Repair: Repair process for joining of fragmented chromosomes is not very accurate in micronuclei and fusion of unwanted chromosome fragments can occur. This can lead to defects in cell proliferation, loss of gene functions and formation of novel oncogene proteins. As detected in cases of pancreatic cancers in which, chromosome 18 and chromosome 12 are most affected [12].
Micronuclei model of chromothripsis has also shown involvement in neochromosome generation. Neochromosomes are functional chromosomes formed by joining of DNA fragments of other normal chromosomes [12]. Involvement of Neochromosomes in cancer is known since 1950 and commonly observed in certain types of lipocarcinomas.
It has been shown that telomere crisis derived kataegis is involved in certain breast cancers. Base substitutions of C>T, C>G and C>A at TpCpX trinucleotides have been observed in various breast cancer samples which is indicative of the same [13].
3. Conclusion and Future Perspective
We have presented a short overview of some the latest discoveries in chromothripsis in cancer that have led to breakthroughs in providing novel insights into cancer genetics. The mechanisms of chromothripsis are discussed, that explain the role of nuclear and somatic genomes in cancer. It shows that formation of micronuclei due to mis-segregation at anaphase stage or telomere loss due to mutated cell cycle checkpoints can lead to tumorigenesis. We have also touched upon the role of nuclear envelope degradation in micronuclei model of chromothripis. This can be further used to study if mitochondrial genome also has the potential to cause cancer by fusing with primary nucleus after degradation of nuclear envelope. Kataegis, a phenomenon of clustering of base substitutions have also been shown to occur due to telomere crisis model of chromothripsis and cause cancer development. Chromothripsis related chromosome rearrangement is an early event in carcinogenesis. These findings can be extended for prognosis and detection of cancer at an early stage and further used in therapeutics.
Acknowledgement
The author (Ishita Agrawal) greatly appreciates the postgraduate fellowship award by the Department of Biotechnology (DBT), Government of India.
References
[1] Cortés-Ciriano, I., Lee, J.JK., Xi, R. et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet 52, 331–341 (2020), DOI: https://doi.org/10.1038/s41588-019-0576-7.
[2] Leibowitz, M. L., Zhang, C.-Z. & Pellman, D. Chromothripsis: a new mechanism for rapid karyotype evolution. Annu. Rev. Genet. 49, 183–211 (2015), DOI: https://doi.org/10.1146/annurev-genet-120213-092228.
[3] Notta, F. et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 538, 378–382 (2016), DOI: https://doi.org/10.1038/nature19823.
[4] Nazaryan-Petersen, L., Bjerregaard, V. A., Nielsen, F. C., Tommerup, N., & Tümer, Z. Chromothripsis and DNA Repair Disorders. Journal of clinical medicine, 9(3), 613 (2020), DOI: https://doi.org/10.3390/jcm9030613.
[5] Shorokhova, M.; Nikolsky, N.; Grinchuk, T. Chromothripsis—Explosion in Genetic Science. Cells. 10, 1102 (2021), DOI: https://doi.org/10.3390/cells10051102.
[6] Jones, M.J. and P.V. Jallepalli, Chromothripsis: chromosomes in crisis. Dev Cell. 23(5), 908-17 (2012), DOI: https://doi.org/ 10.1016/j.devcel.2012.10.010.
[7] Rode, A., et al., Chromothripsis in cancer cells: An update. Int J Cancer. 138(10), 2322-33 (2016), DOI: https://doi.org/ 10.1002/ijc.29888.
[8] Liu, S., Kwon, M., Mannino, M. et al. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature. 561, 551–555 (2018), DOI: https://doi.org/10.1038/s41586-018-0534-z
[9] Hatch, E.M. and M.W. Hetzer, Linking Micronuclei to Chromosome Fragmentation. Cell. 161(7), 1502-4 (2015), DOI: https://doi.org/ 10.1016/j.cell.2015.06.005.
[10] Zhang, CZ., Spektor, A., Cornils, H. et al. Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015), DOI: https://doi.org/10.1038/nature14493.
[11] Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 163(7):1641-1654 (2015), DOI: https://doi.org/10.1016/j.cell.2015.11.054.
[12] Luijten, M.N.H., J.X.T. Lee, and K.C. Crasta, Mutational game changer: Chromothripsis and its emerging relevance to cancer. Mutation Research/Reviews in Mutation Research. 777, 29-51 (2018), DOI: https://doi.org/10.1016/j.mrrev.2018.06.004.
[13] Nik-Zainal, S., et al., Mutational processes molding the genomes of 21 breast cancers. Cell. 149(5), 979-93 (2012), DOI: https://doi.org/ 10.1016/j.cell.2012.04.024.
Rights and Permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third-party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ Creative Commons This is an open access article distributed under the terms of the Creative Commons CC BY license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. You are not required to obtain permission to reuse this article. To request permission for a type of use not listed, please contact Biotechnology Kiosk.