Electron Microscopy for Structural Determination and Analysis of Viruses
Sandeep Kumar Yadav
Department of Genomic Medicine, University of Texas, MD Anderson Cancer Center, Houston, TX, USA
Abstract
Viruses are known to be associated with large-scale, dynamic conformational changes that take place to facilitate cell entry and genome delivery. It is also known that a replication machinery is involved in the advanced stage of the infectious cycle that enables to read and synthesize nucleic acid strands. This process results in the generation of new copies of genetic material. In this process, the function of structural proteins helps to assemble and package the appropriate contents to produce new infectious particles. Lately, there has been a great deal of research interest on structural elucidation of these events. This interest is primarily driven by the significance of virus structural identification, which helps understand these processes and also their inhibition by antiviral agents such as neutralizing antibodies and drugs. To this end, the development of electron microscopy (EM) techniques for studies in virology has played a major role for structural determination and analysis of viruses. In this mini-review, we have highlighted some of the latest developments in this field. We have briefly described important role of EM in virology. We have also discussed notable application examples of EM in elucidating various virus structures to gain insights into identification of deadly pathogens and other infectious agents and outbreaks along with anti-viral developmental strategies.
Keywords: Electron microscopy, cryo-electron microscopy, virus, structure, antiviral therapy, genome delivery, cells, replication.
DOI: https://doi.org/10.37756/bk.22.4.2.2
Article type: Mini-Review
Received: December 10, 2021
Revised: January 05, 2022
Accepted: January 18, 2022
Please cite this article as: Yadav SK, Electron Microscopy for Structural Determination and Analysis of Viruses, Biotechnol. kiosk, Vol 4, Issue 2, PP: 12-25 (2022); DOI: https://doi.org/10.37756/bk.22.4.2.2
*Corresponding author: Dr. Sandeep Kumar Yadav
E-mail address: sandeepyadav.bhu@gmail.com
Introduction
The Role of Electron Microscopy in Virology
Viruses are known to be extremely diverse in shape and size. Over the years, numerous studies on viruses have shed light on their evolution that has led to a limited number of viral classes or lineages [1-6]. In addition, virologists have used the term virosphere to describe general features of viruses that not only include the space where viruses are found but also the space they influence that can extend their impact on the environment. This scenario explains the complexity of the interactions involved in the viral assembly mechanisms that are restricted to two general pathways. The co-assembly of capsid proteins and single-stranded nucleic acids is one such pathway. The other pathway involves a sequential mechanism in which scaffolding-mediated capsid precursor assembly is followed by genome packaging [7-12].
In order to prevent viral disease along with virus control and establishing efficient and reliable virus diagnosis, it is critical that studies on the biology of viruses and the etiology of virus disease are conducted. To address these issues, researchers have shown that knowledge of the atomic resolution structures of viruses can be a powerful tool for vaccine discovery and design to mitigate the challenges of global pandemics caused by deadly viruses [13-14]. To this end, the role of electron microscopy (EM) in virology is believed to be pivotal that can be leveraged to identify the causative agents of infectious diseases. EM has been shown to be an essential tool in the detection and analysis of virus replication. For example, the technique has been proven to be important to diagnose pathogens and in testing to identify microorganisms. New EM methods and ongoing technical improvements have been shown to cover a broad range of applications that have allowed in-depth investigation of viral impact on not only the host but also the environment. It is now possible to conduct such investigations close to atomic resolution with the latest cryo-EM based methods. Further, it has been shown that in combination with bioinformatics, the transition from two-dimensional ‘2D’ imaging to three-dimensional ‘3D’ remodeling can be accomplished. This allows structural and functional analyses that advance our fundamental understanding of the diversity in virus structure and lifestyle. Especially, EM in combination with advanced techniques such as confocal laser scanning microscopy can be leveraged for live imaging of cells and tissues with high-resolution analysis [7, 15-17].
EM is advantageous over conventional light microscopy for the reason of the technique’s capability to analyze small size of virus particles that cannot be visualized by light microscopy based techniques [18-21]. Researchers have shown the versatility of EM to resolve 3D structures of individual viral proteins and whole virions in multiple functional states that include cells at different stages of infection. This makes EM as a very popular technique that can be employed to identify and analyze unknown emerging viruses even when there are no primers, antibodies or probes available. Thus, EM is considered extremely advantageous because of a generic approach that it follows that offers the potential to detect all viral particles present in a sample [22, 23].
Preparation and Analysis of Virus-Infected Cells for Electron Microscopy
It is very important that the ultrastructure is preserved in a state that is as close as possible to a snapshot of the living state. Accordingly, it is suggested that the preparation of cells for EM should follow the major goal of preserving the cell structure that is visualized at the resolution limit of the electron microscope. To this end, researchers have shown several methods and standard recipes. These methods can be frequently modified when addressing particular biological questions [22, 23].
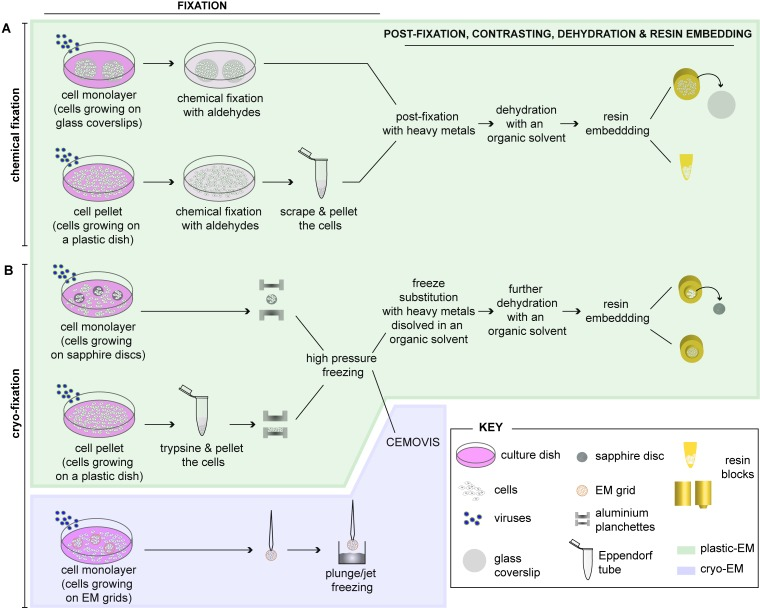
Figure 1: [A-B] Schematic depiction highlighting different methods for preparing virus-infected cells for electron microscopy [Source: Viruses (2015)].
Figure 1 shows the standard method to prepare cells for routine EM that involves fixation, embedding and sectioning [23]. As Figure 1 shows, adherent cells can be either chemically fixed with aldehydes or fast frozen by high-pressure freezing (HPF) or plunge/jet freezing. Subsequently, infected cells can be further processed as a cell monolayer (e.g. grown on glass coverslips) or be pelleted prior to further processing for EM after chemical fixation. Post-fixation can be done with heavy metals using osmium tetroxide (OsO4) and uranyl acetate (UA). This results in sample dehydration with increasing concentrations of an organic solvent using ethanol or acetone. This is followed by embedding into a plastic resin (plastic-EM; highlighted in green) (Figure 1). It is also suggested that coverslips are removed from the polymerized resin block. This can be done by successive immersions in liquid nitrogen and hot water. Further, they must be grown as monolayers on sapphire discs for rapid immobilization of cells by HPF. This can be achieved by clamping in-between two aluminum planchettes and subsequently loading into a HPF machine for rapid freezing [23]. Alternatively, this can be done by directly placing cell pellets into the aluminum planchettes or into capillary tubes for freezing. Frozen cells can then be subjected to freeze substitution (FS), dehydration with an organic solvent and resin embedding (plastic-EM; highlighted in green in Figure 1). Further, as an alternative approach, high-pressure frozen cell pellets can be analyzed by CEMOVIS (cryo-electron microscopy of vitreous sections) (cryo-EM, highlighted in violet in Figure 1). Cells growing on EM grids can then be plunge/jet frozen and analyzed directly by cryo-EM. Both these cryo-EM approaches do not require further processing of the cells and allow visualizing cells in their closest-to-native status [23].
With respect to analyzing virus-infected cells, the main research focus has been on visualization comprising different steps of the viral life cycle that include attachment, entry, replication, assembly and egress along with the study of the ultrastructure of the infected cell. To this end, an important goal is to understand virus-induced modifications of a targeted subcellular organelle. This is to elucidate not only the function of different viral proteins, but also aimed at designing novel antiviral drugs and eventually vaccines [24].
Several analytical approaches have been developed to gain 3D information of cellular structures by EM. Researchers have shown the possibility to unravel the 3-D architecture of cellular organelles and even complete virus-infected cells with the latest advances and the ongoing implementation of novel EM techniques (Figure 2) [23]. For example, negative stain transmission electron microscopy (TEM) is frequently considered for imaging microbial samples especially in diagnostic virology. However, there is a challenge in negative-stain TEM that requires an adequate concentration of bacterial cells or virus particles. To address this issue, researchers have grown microbes to a high tire and/or concentrated by centrifugation. However, such a process is somewhat difficult to do with patient specimens or agents that are not easy to culture. There has been some progress in filtration techniques that have shown that both TEM and scanning electron microscope (SEM) identification of viruses can be carried out with as little as 5000 total particles per sample. Studies have also shown that SEM can be employed to study morphological features of isolated organisms for diagnosis purposes. In such cases, biological specimens are required to be coated using thin film evaporation or sputtering of carbon or metal in a vacuum coater to produce an electrically conductive surface for SEM [24].
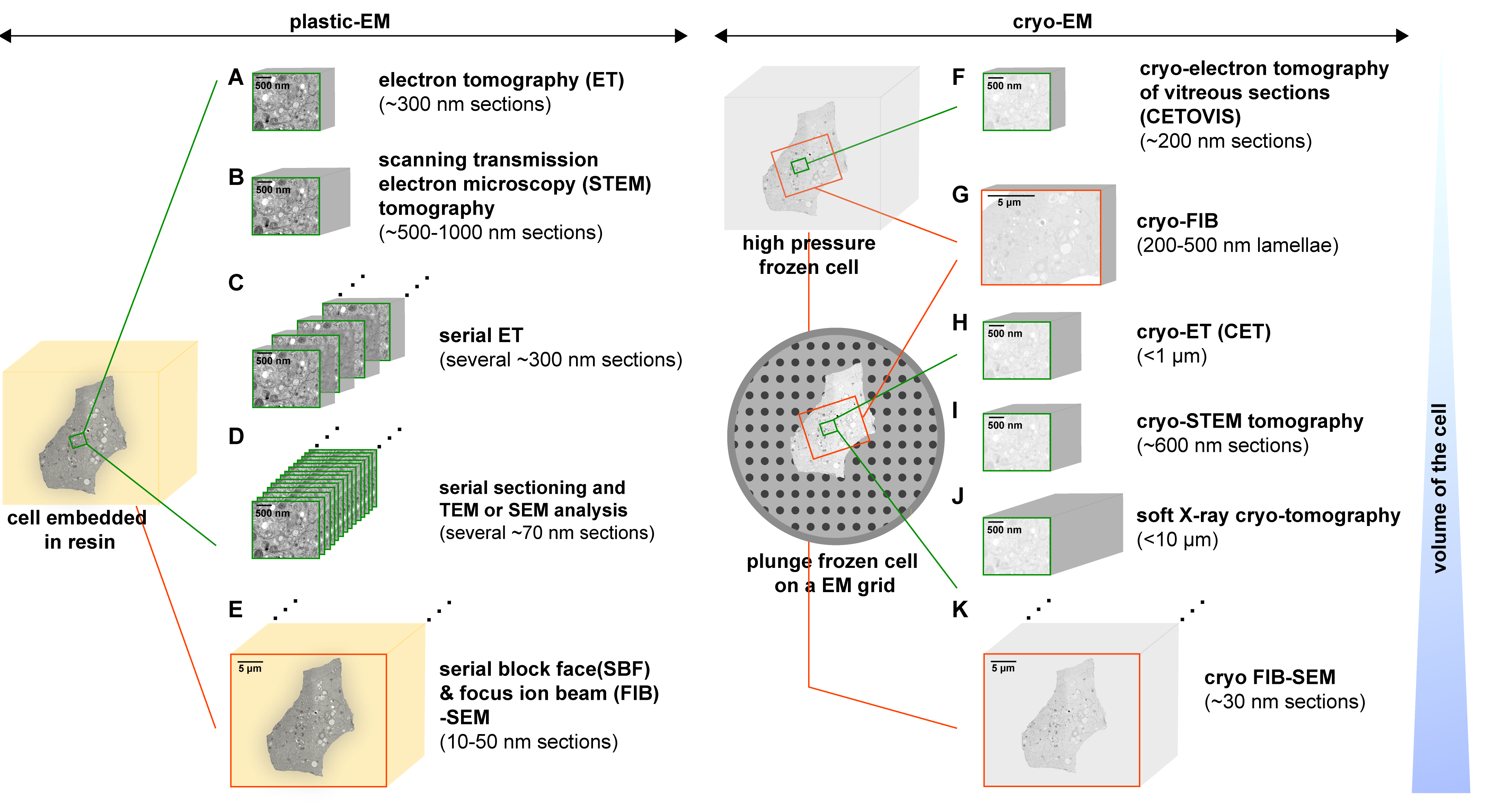
Figure 2: An overview of the 3D-EM methods that show various approaches to analyze the architecture of virus-infected cells. This is achieved by employing plastic-EM (A-E) or cryo-EM (F-K) [Source: Viruses (2015)].
Lately, cryo-EM has emerged as a popular technique for virus detection and analysis of specimens that have not been stained or fixed in any way. Cryo-EM and cryo-electron tomography (cryo-ET) (Figure 2) are the latest EM technologies that are revolutionizing the field of structural biology. Recent research has shown the huge promise of cryo-EM to determine high-resolution structures of many viral assemblies as well as those of assembly intermediates. This paves the way for further development and redesign of virus-based platforms for advanced biomedical and biotechnological applications [25, 26].
In the following sections, we have described some of the important applications of electron microscopy in detection, identification and analysis of complex viruses.
Investigating Novel Viruses from Mud Crab Scylla paramamosain with Multiple Infections by Cryo-Electron Microscopy
Identification of deadly pathogen is considered very important for early intervention in unknown infectious outbreaks. Such early identifications are especially recommended for pathogens that can cause dual or multiple infections. One example is the mud crab Scylla paramamosain, which is an economically important aquaculture species that are found in China, India, Australia, and many other countries. The problem is however, the prevalence of sleeping disease (SD) that is found in this species that results in significant economic losses. Previous studies involving molecular investigations were conducted that showed two major viral pathogens including mud crab reovirus (MCRV) and mud crab dicistrovirus (MCDV) with diameters of 72 and 30 nm, respectively. MCDV has been reported to belong to the family Dicistroviridae, which contains three genera involving Cripavirus, Triatovirus, and Aparavirus [27-31].
SD in crabs with multiple infections is known to affect aquaculture worldwide in addition to potential outbreaks in human population. Researchers addressed these issues in a recent study and they reported discovery and identification of a novel virus in mud crabs with multiple infections by employing cryo-EM. Such virus structural analysis was not previously done by molecular, immune, or traditional electron microscopy methods. In this study, researchers showed high-resolution structures of pathogenic viruses that provided new ways for a molecular understanding and developing new disease prevention methods. They showed that the 3D structure of the mud crab tombus-like virus (MCTV) and mud crab dicistrovirus (MCDV) could assist the development of antiviral inhibitors. The identification of a novel virus in multiple infections demonstrated the applicability of this strategy for investigating multiple infectious outbreaks including humans and other animals [32].
Single-particle cryo-EM was employed to investigate viruses associated with SD following novel antiviral developmental strategies (Figure 3) [32]. The results revealed the structure of mud crab dicistrovirus (MCDV) along with identifying a novel mud crab tombus-like virus (MCTV) that was not previously detected using molecular biology methods. Moreover, the structure of MCDV at a 3.5-Å resolution identified three major capsid proteins (VP1 to VP3) that were found organized into a pseudo-T3 icosahedral capsid, which confirmed the existence of VP4. The researchers also made an unusual observation on MCDV VP3 that contained a long C-terminal region, which formed a novel protrusion that was not observed in other prior studies on dicistrovirus. These results revealed that MCDV can release its genome via conformation changes of the protrusions when viral mixtures were heated. Also, the structure of MCTV at a 3.3-Å resolution revealed a T 3 icosahedral capsid with common features of both tombusviruses and nodaviruses [32].
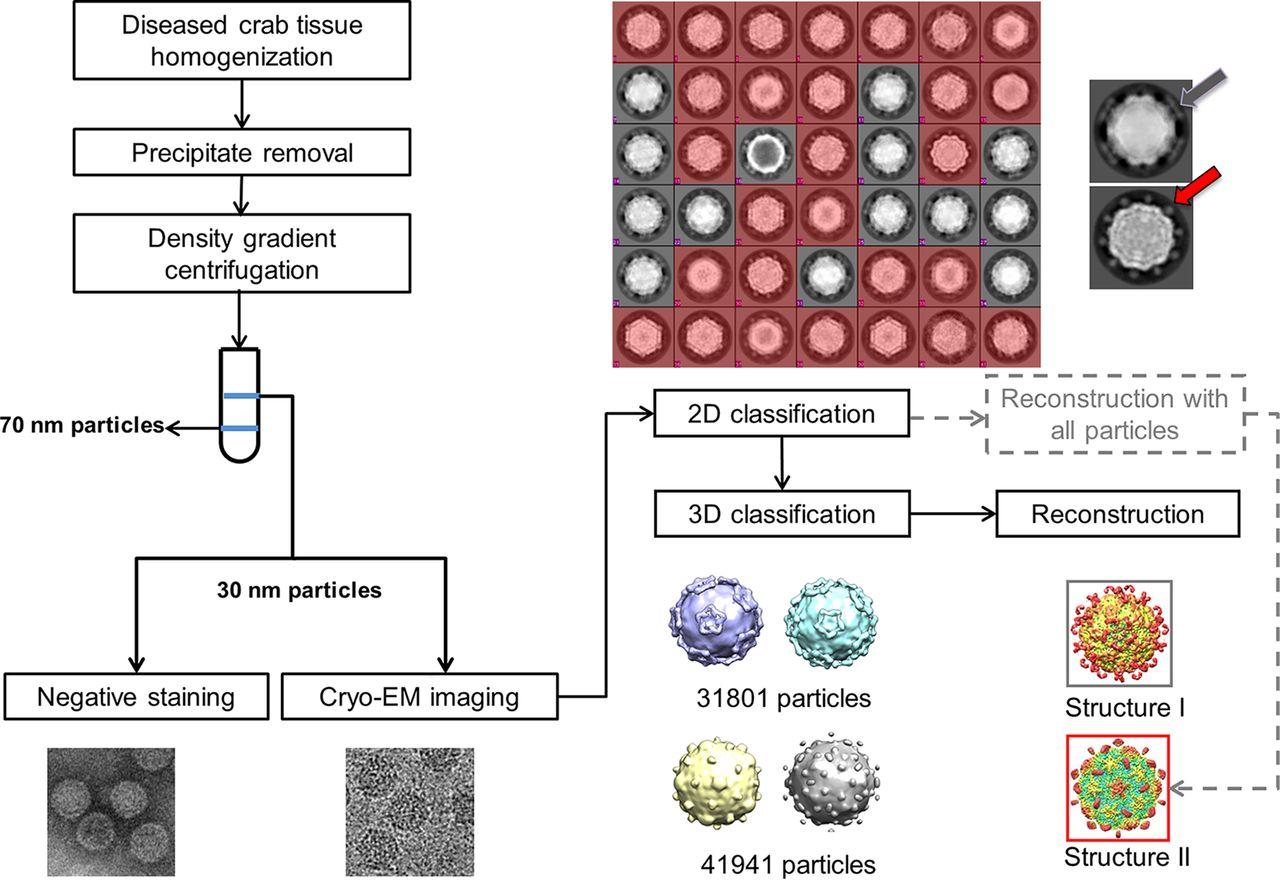
Figure 3: Flow chart is shown for sample isolation and data processing that revealed two kinds of virus based on the shape of protrusions including one (colored red) that showed dot-like protrusions (red arrow), and the other (gray) showed lamina-shaped protrusions (gray arrow). These results confirmed the presence of two kinds of virus with different protrusions [Source: Journal of Virology (2019)].
Identification and Analysis of Viral Infection of Tissues in COVID-19 by Transmission Electron Microscopy
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, causing coronavirus disease 2019 (COVID-19) has devastated the global population that resulted in millions of death worldwide. To gain insights into the novel virus, there have been intense investigations into the pathogenesis of this disease. With the emergence of SARS-CoV-2, one of the research areas that has received great attention is the use of EM to help identify virally infected cells and uncover the pathogenesis of this disease. To this end, several articles have used EM that have proposed direct evidence of infection of the kidney and other tissues by SARS-CoV-2. These reports have triggered speculation that the morbidity and mortality of COVID-19 have resulted from direct infection of tissues throughout the body. These studies have indicated that the identification of pathogens/viruses by electron microscopy help to establish critical information that is needed to better understand the biology of the disease and achieve effective treatments for this and future pandemics [33-39].
In a recent relevant study, researchers employed TEM for the identification of novel viruses including SARS. They showed effectiveness of TEM for the identification of COVID-19 in tissues and morphologic features of the viral particles that were consistent with the prior knowledge of the virus. This study focused on understanding size and uniformity, formation of higher order structures including aggregates/arrays/inclusions, the absence or presence of a clearly discernible membrane, and also the qualities of nucleocapsid and peplomers/spikes electron densities [40].
By employing TEM, researchers investigated the SARS-CoV2 viral particles that were shown to be with an average diameter of 64 nm (range 56-75 nm) (Figure 4) [40]. It was also shown that tissue preservation was critical because poor preservation for autopsy material could often compromise objective interpretation of electron micrographs and the ability to conclusively identify viral particles. Location of viral particles was also studied by TEM that conformed to the known biology of viral replication. This provided strong supporting evidence that was required when attempting to identify viral particles in tissues with suboptimal preservation, necrosis and autolysis in order to differentiate these particles from normal cellular structures. As shown in Figure 4, coronavirus particles were found inside the cisternae of the ER-Golgi and secretory compartment, as well as outside of cells [40].
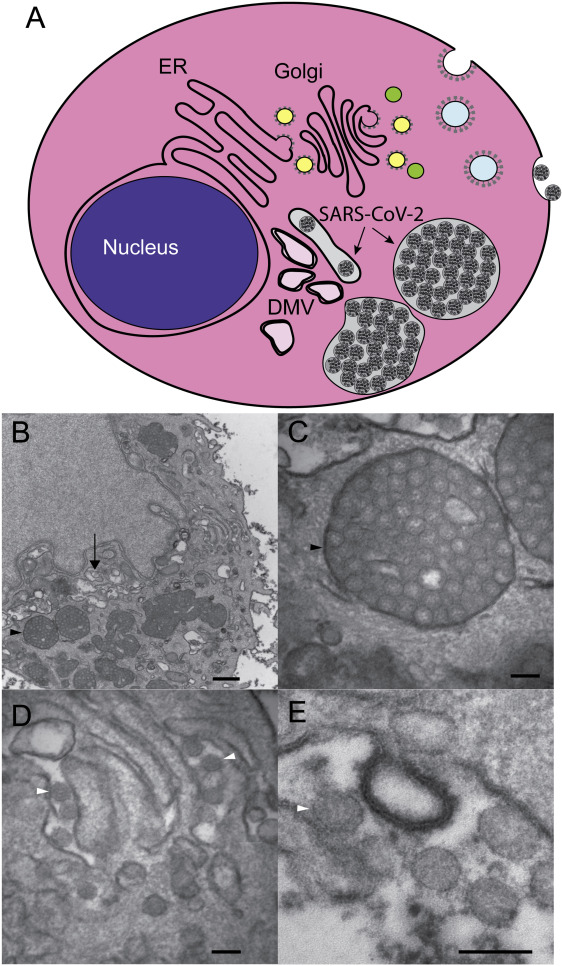
Figure 4: [A-E] Schematic depiction of cell demonstrating structures that are associated with the investigations of coronavirus infection in cells by transmission electron microscope (TEM). This shows double-membrane vesicles (DMV) that are found near the nucleus and represent the site of viral genome replication. Coronavirus particles bud into the cisternae of the ER/Golgi and they accumulate in cytoplasmic vesicles that fuse with the plasma membrane and release virus particles into the extracellular space. Typical TEM images are shown for a SARS-CoV-2 infected HBEC3-KT cell with perinuclear DMV (arrow) and enlarged vesicles (black arrowhead) filled with viral particles (bar = 500 nm) [Source: The American Journal of Pathology (2020)].
Scanning Transmission Electron Microscopy (STEM) Detector for Virus Quantitation
Previous studies have shown that virus quantitation is an important factor when studying the environmental impact of viruses, or virus impact on a specific host. To this end, an accurate method for the quantitation of virus particles is considered vital and therefore, the research in virology has been focused on developing a universally accepted method that can be adopted by the scientific community. It is believed that an accurate method for the quantitation of virus particles is very useful. However, a universally accepted method has not been adopted by the scientific community so far. To address the issue, researchers have employed direct EM quantitation, which is considered a valuable technique due to the reason that it provides enumeration of all virus particles including infectious non-infectious [41-44].
In a recent study, researchers have demonstrated a method for accurate quantitation of virus particles based on electron microscopy quantitation that provides direct morphology information and counts of all viral particles, whether or not they are infectious [45]. In the past, EM negative stain quantitation methods were cited as inaccurate, non-reproducible, and that were too high to be useful. In this study, researchers improved accuracy and reproducibility with detection limits by employing a method termed Scanning Transmission Electron Microscopy – Virus Quantitation (STEM-VQ), which simplified sample preparation and that used a high throughput STEM detector in a SEM coupled with commercially available software. They demonstrated STEM-VQ with an alphavirus stock preparation to present the method’s accuracy and reproducibility, including a comparison of STEM-VQ to viral plaque assay and the ViroCyt Virus Counter (Figure 5) [45].
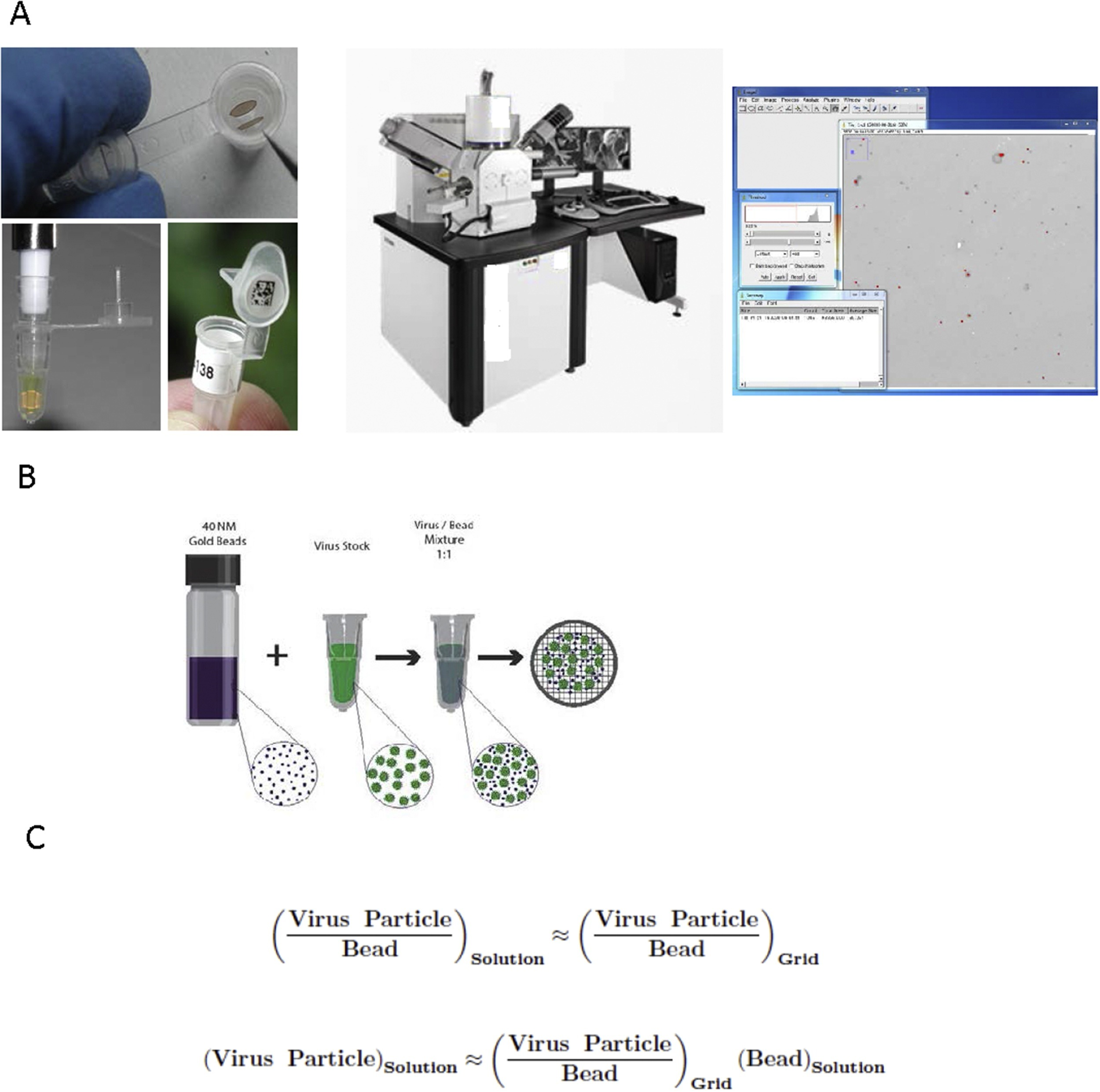
Figure 5: [A-C] An overview of the employed STEM-VQ method is shown with the three major phases that are needed for determining particle concentration (left) sample preparation using mPrep/g system, (middle) STEM imaging in the SEM and (right) particle counting using image. A mixture of a known concentration of gold beads is also shown with an unknown concentration of virus stock and a formula is shown that is used to calculate the number of unknown viral particles based on the known concentration of gold beads and the virus-gold ratio [Source: Journal of Virological Methods (2017)].
Conclusion
In this mini-review, we have highlighted advanced electron microscopy (EM) based methods that have proven to be immensely promising in elucidating and deeper understanding of the structures of complex and deadly viruses. We have presented some notable examples of EM based methods that have been shown to be capable of detecting and analyzing viruses such as SARS-CoV-2. It is believed that these investigations are of hugely important because a deeper understanding of virus structural morphology could help pave the way to major breakthroughs in the future in the areas of new therapeutics, such as antibodies and drugs and eventually highly effective and safe vaccines. It is a multidisciplinary research field that involves thin film technologists and electron microcopy experts along with biotechnologists and virologists, to name a few. It is anticipated that future studies will include combination of different EM techniques to elucidate ultra-small structural features of viruses that will help understand different stages of infectious cycle along with new insights into virosphere. This will eventually help prevent future outbreaks and pandemics.
References
[1] Earl LA and Subramaniam S , Cryo-EM of viruses and vaccine design. Proc. Natl. Acad. Sci. USA 2016, 113 (32) 8903-8905, DOI: https://doi.org/10.1073/pnas.1609721113
[2] Mettenleiter TC, The first virus hunters. Advances in Virus Research 2017, eds M. Beer and D. Höper (Burlington: Academic Press), 99:1–16, DOI: https://doi.org/10.1016/bs.aivir.2017.07.005
[3] Böttcher B, Wynne SA, and Crowther RA, Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 1997, 386:88–91, DOI: https://doi.org/10.1038/386088a0
[4] Brenner S, and Horne RW, A negative staining method for high resolution electron microscopy of viruses. Biochim. Biophys. Acta 1959, 34:103–110, DOI: https://doi.org/10.1016/0006-3002(59)90237-9
[5] Zamora M, Méndez-López E, Agirrezabala X, Cuesta R, Lavin JL, Sánchez Pina MA, et al., Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Sci. Adv 2017, 3:1–7, DOI: https://doi.org/10.1126/sciadv.aao2182
[6] Zhang W, Olson NH, Baker TS, Faulkner L, Agbandje-McKenna M, Boulton MI, et al., Structure of the Maize streak virus geminate particle. Virology 2001, 279:471–477, DOI: https://doi.org/10.1006/viro.2000.0739
[7] Richert-Pöggeler KR, Franzke K, Hipp K and Kleespies RG, Electron Microscopy Methods for Virus Diagnosis and High Resolution Analysis of Viruses. Front. Microbiol 2019, 9:3255, DOI: https://doi.org/10.3389/fmicb.2018.03255
[8] Valle M, “Structural homology between nucleoproteins of ssRNA viruses,” in Subcellular Biochemstry 88. Virus Protein and Nucleoprotein Complexes, eds J. Harris and D. Bhella. Singapore: Springer 2018, 129–145, DOI: https://doi.org/10.1007/978-981-10-8456-0_6
[9] Romette JL, Prat CM, Gould EA, de Lamballerie X, Charrel R, Coutard B, et al., The European Virus Archive goes global: a growing resource for research. Antiviral Res 2018, 158:127–134, DOI: https://doi.org/10.1016/j.antiviral.2018.07.017
[10] Clare DK, Pechnikova EV, Skurat EV, Makarov VV, Sokolova OS, Solovyev AG, et al., Novel inter-subunit contacts in barley stripe mosaic virus revealed by cryo-electron microscopy. Structure 2015, 23:1815–1826, DOI: https://doi.org/10.1016/j.str.2015.06.028
[11] Koonin EV, Dolja VV, and Krupovic M, Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 2015, 479–480, 2–25, DOI: https://doi.org/10.1016/j.virol.2015.02.039
[12] Ghoshal K, Theilmann J, Reade R, Maghodia A, and Rochon D, Encapsidation of host RNAs by cucumber necrosis virus coat protein during both agroinfiltration and infection. J. Virol 2015, 89:10748–10761, DOI: https://doi.org/10.1128/jvi.01466-15
[13] Agirrezabala X, Méndez-López E, Lasso G, Sánchez-Pina MA, Aranda M, and Valle M, The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses. Elife 2015, 4:e11795, DOI: https://doi.org/10.7554/eLife.11795
[14] Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JLS, et al. 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science 2015, 348:1147–1151, DOI: https://doi.org/10.1126/science.aab1576
[15] Biel SS, Madeley D, Diagnostic virology—The need for electron microscopy: A discussion paper. J. Clin. Virol 2001, 22:1–9, DOI: https://doi.org/10.1016/S1386-6532(01)00151-2
[16] Roingeard P, Viral detection by electron microscopy: Past, present and future. Biol. Cell 2008, 100:491–501, DOI: https://doi.org/10.1042/BC20070173
[17] Hazelton PR, Gelderblom HR, Electron microscopy for rapid diagnosis of infectious agents in emergent situations. Emerg. Infect. Dis 2003, 9:294–303, DOI: https://doi.org/10.3201/eid0903.020327
[18] Zhou ZH, Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr. Opin. Struct. Biol 2008, 18:218–228, DOI: https://doi.org/10.1016/j.sbi.2008.03.004
[19] Cheng Y, Walz T, The advent of near-atomic resolution in single-particle electron microscopy. Annu. Rev. Biochem 2009, 78:723–742, DOI: https://doi.org/10.1146/annurev.biochem.78.070507.140543
[20] Wolf M, Garcea RL, Grigorieff N, Harrison SC, Subunit interactions in bovine papillomavirus. Proc. Natl. Acad. Sci. USA, 2010, 107:6298–6303, DOI: https://doi.org/10.1073/pnas.0914604107
[21] Bharat TA, Davey NE, Ulbrich P, Riches JD, de Marco A, Rumlova M, Sachse C, Ruml T, Briggs JA, Structure of the immature retroviral capsid at 8 A resolution by cryo-electron microscopy. Nature 2012, 487:385–389, DOI: https://doi.org/10.1038/nature11169
[22] Risco C, de Castro IF, Sanz-Sanchez L, Narayan K, Grandinetti G, Subramaniam S, Three-dimensional imaging of viral infections. Annu. Rev. Virol 2014, 1:453–473, DOI: https://doi.org/10.1146/annurev-virology-031413-085351
[23] Romero-Brey I, Bartenschlager R. Viral Infection at High Magnification: 3D Electron Microscopy Methods to Analyze the Architecture of Infected Cells. Viruses 2015, 7(12):6316-45, DOI: https://doi.org/10.3390/v7122940
[24] Golding C, Lamboo L, Beniac D et al., The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci Rep 2016, 6:26516, DOI: https://doi.org/10.1038/srep26516
[25] Luque D, Castón JR, Cryo-electron microscopy for the study of virus assembly. Nat Chem Biol 2020, 16(3):231-239. DOI: https://doi.org/10.1038/s41589-020-0477-1
[26] Roingeard P, Raynal PI, Eymieux S, Blanchard E, Virus detection by transmission electron microscopy: Still useful for diagnosis and a plus for biosafety. Rev Med Virol. 2019, 29(1):e2019, DOI: https://doi.org/10.1002/rmv.2019
[27] Huang ZW, Deng XX, Li YY, Su HJ, Li KP, Guo ZX, Zheng PR, Xu HD, He JG, Zhang QF, Weng SP, Structural insights into the classification of mud crab reovirus. Virus Res 2012, 166:116–120, DOI: https://doi.org/10.1016/j.virusres.2012.02.025
[28] Zhang R, He J, Su H, Dong C, Guo Z, Ou Y, Deng X, Weng S, Identification of the structural proteins of VP1 and VP2 of a novel mud crab dicistrovirus. J Virol Methods 2011, 171:323–328. DOI: https://doi.org/10.1016/j.jviromet.2010.09.010
[29] Guo ZX, He JG, Xu HD, Weng SP, Pathogenicity and complete genome sequence analysis of the mud crab dicistrovirus-1. Virus Res 2013, 171:8–14. DOI: https://doi.org/10.1016/j.virusres.2012.10.002
[30] Deng XX, Lu L, Ou YJ, Su HJ, Li G, Guo ZX, Zhang R, Zheng PR, Chen YG, He JG, Weng SP, Sequence analysis of 12 genome segments of mud crab reovirus (MCRV). Virology 2012, 422:185–194. DOI: https://doi.org/10.1016/j.virol.2011.09.029
[31] Le Gall O, Christian P, Fauquet CM, King AMQ, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE, Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch Virol 2008, 153:715–727, DOI: https://doi.org/10.1007/s00705-008-0041-x
[32] Gao Y, Liu S, Huang J, Wang Q, Li K, He J, He J, Weng S, Zhang Q, Cryo-electron Microscopy Structures of Novel Viruses from Mud Crab Scylla paramamosain with Multiple Infections. J Virol 2019, 21;93(7):e02255-18, DOI: https://doi.org/10.1128/JVI.02255-18
[33] Bradley BT, Maioli H, Johnston R, Chaudhry I, Fink SL, Xu H, Najafian B, Deutsch G, Lacy JM, Williams T, Yarid N, Marshall DA, Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet 2020, 1;396(10247):320-332, DOI: https://doi.org/10.1016/S0140-6736(20)31305-2
[34] Farkash EA, Wilson AM, Jentzen JM, Ultrastructural Evidence for Direct Renal Infection with SARS-CoV-2. J Am Soc Nephrol 2020, 31(8):1683-1687, DOI: https://doi.org/10.1681/ASN.2020040432
[35] Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, Li WW, Li VW, Mentzer SJ, Jonigk D, Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N Engl J Med 2020, 9;383(2):120-128, DOI: https://doi.org/10.1056/NEJMoa2015432
[36] Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F, Moch H, Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 2;395(10234):1417-1418, DOI: https://doi.org/10.1016/S0140-6736(20)30937-5
[37] Algarroba GN, Rekawek P, Vahanian SA, Khullar P, Palaia T, Peltier MR, Chavez MR, Vintzileos AM, Visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am J Obstet Gynecol 2020, 223(2):275-278, DOI: https://doi.org/10.1016/j.ajog.2020.05.023
[38] Pesaresi M, Pirani F, Tagliabracci A, Valsecchi M, Procopio AD, Busardò FP, Graciotti L, SARS-CoV-2 identification in lungs, heart and kidney specimens by transmission and scanning electron microscopy. Eur Rev Med Pharmacol Sci 2020, 24(9):5186-5188, DOI: https://doi.org/10.26355/eurrev_202005_21217
[39] Tavazzi G, Pellegrini C, Maurelli M, Belliato M, Sciutti F, Bottazzi A, Sepe PA, Resasco T, Camporotondo R, Bruno R, Baldanti F, Paolucci S, Pelenghi S, Iotti GA, Mojoli F, Arbustini E, Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur J Heart Fail 2020, 22(5):911-915, DOI: https://doi.org/10.1002/ejhf.1828.
[40] Akilesh S, Nicosia RF, Alpers CE, Tretiakova M, Hsiang TY, Gale M Jr, Smith KD, Characterizing Viral Infection by Electron Microscopy: Lessons from the Coronavirus Disease 2019 Pandemic. Am J Pathol 2021, 191(2):222-227, DOI: https://doi.org/10.1016/j.ajpath.2020.11.003
[41] Ferris MM, Stoffel CL, Maurer TT, Rowlen KL, Quantitative intercomparison of transmission electron microscopy, flow cytometry, and epifluorescence microscopy for nanometric particle analysis. Anal. Biochem 2002, 304:249-256, DOI: https://doi.org/10.1006/abio.2002.5616
[42] Malenovska H, Virus quantitation by transmission electron microscopy, TCID(5)(0), and the role of timing virus harvesting: a case study of three animal viruses. Virol. Methods 2013, 191:136-140, DOI: https://doi.org/10.1016/j.jviromet.2013.04.008
[43] Rossi CA et al., Evaluation of ViroCyt(R) Virus Counter for rapid filovirus quantitation. Viruses 2015, 7:857-872, DOI: https://doi.org/10.3390/v7030857
[44] Monninger MK, et al., Preparation of viral samples within biocontainment for ultrastructural analysis: utilization of an innovative processing capsule for negative staining. J. Virol. Methods 2016, 238:70-76, DOI: https://doi.org/10.1016/j.jviromet.2016.10.005
[45] Blancett CD, Fetterer DP, Koistinen KA, Morazzani EM, Monninger MK, Piper AE, et al., Accurate virus quantitation using a Scanning Transmission Electron Microscopy (STEM) detector in a scanning electron microscope. J. Virol. Methods 2017, 248:136–144, DOI: https://doi.org/10.1016/j.jviromet.2017.06.014