
Article type: Mini Review
Received: June 14, 2020
Revised: July 01, 2020
Accepted: July 15, 2020
Citation: Singh P and Singh S; Effective countermeasure for Filovirus infections: focusing monoclonal antibodies as passive prophylaxis shield against Ebola Virus disease, Biotechnology Kiosk, Vol 2, Issue 8, PP: 18-35 (2020); DOI: https://doi.org/10.37756/bk.20.2.8.2
Abstract
In 2020 the pandemic of COVID 19 by SARS-COV 2 infected more than 27 million with more than 875,000 deaths. Present day world is more compact with quick mode of transport between far locations, this makes spread of new viral infections at alarming rates. Similarly, Filovirus is a family of extremely dangerous Marburgvirus and Ebolavirus with up to 90% mortality rate. Since first Filovirus was discovered in 1967, many outbreaks were reported from African countries. Increased number of infected human population in other outbreaks in 2014 – 2017 poses a question of our understanding of Filovirus reservoirs. We are beginning to understand the relation between virus and ecological agents and their role in the spread of disease, but it is still a long road ahead. To counteract and containment of Filovirus infection, it is utmost requirement to understand the viral life cycle patterns, agents involved and type of circulating strains in different geographical locations. This information will provide the basis to develop viable therapies to counteract future outbreaks. In these outbreaks’ magnitude of population and geographical area affected creates the urgency to generate effective vaccines and prophylactic agents so that mortalities can be controlled during future outbreaks. Therapies are required for pre-infection acute phase and post-infection. Here, we summarize recent advances in immunotherapy strategies that can be used as passive prophylaxis. We focused on development of recent monoclonal antibodies and cocktails that can be used as neutralizing agents or immunotherapy for Ebola infected patients During the pre-outbreak period it is required to vaccinate susceptible populations that will allow limiting the infection and mortalities. Furthermore, during the acute phase to neutralize virus and limiting disease symptoms, passive
prophylaxis mean neutralizing antibodies are required. In the recent past few promising therapies are developed, some of these are on the clinical trial phase. Here we will review these therapies with their advantages in protecting against Filovirus.
Key Words: Pandemic, COVID 19, neutralizing antibodies
Introduction
As most of the world is now connected with air, land, and sea routes, the COVID-19 pandemic will become a classic example of how a single virus can affect the whole world. This pandemic possesses a real threat to human life and the world economy. COVID 19 situation brings the focus of whole scientific community on Corona CoV 2 testing, treatment, and vaccination strategies. We are living in a world with the continuous threat of new disease and infection, here we have focused on available effective countermeasures for Filovirus infections, focusing on monoclonal antibodies.
On May 11, 2017, the Ministry of Public Health of the Democratic Republic of the Congo notified international public health agencies of a cluster of suspected cases of Ebola Virus Disease (EVD) in the Likati health zone of the province of Bas Uélé. Teams from international agencies, including CDC, WHO, MSF (Doctors without Borders), and others, supported the Ministry of Public Health’s epidemiologic, diagnostic, clinical, and communications efforts to respond to the outbreak. The response faced challenging logistical obstacles, including the remoteness of the area and limited services. Mobile diagnostic laboratories provided testing of samples in the affected areas. Following a period of 42 days since the second negative laboratory diagnostic test of the last confirmed patient, WHO declared an end to the outbreak on July 2, 2017. Summarizing the total 8 cases (probable or confirmed) 5 were laboratory confirmed and 4 died (50%) (https://www.cdc.gov/vhf/Ebola/outbreaks/history/chronology.html).
Earlier Ebola Virus Disease (EVD) was assumed to be too lethal to spread to large geographical areas as the disease was associated with high mortality rates of upto 100% cases (CDC, USA 2020). However, after 2013 outbreak primarily in Guinea, Liberia and Sierra Leone with 28,616 human cases, health agencies were forced to make fundamental changes in general view regarding the potential of EVD as a global threat.
The global health community recognizes the urgent need to develop strategies that can be used before, during and after any future Filovirus outbreak. These include development of vaccines and immunotherapy. The information regarding the eco – biology of Filovirus greatly hinder the development of accurate therapies. Even after more than 50 years, our understanding of Filovirus natural reservoirs is very limited. The limited information on natural host and pathology of Filovirus is one of the major causes of the limited development of new therapies.
Here, we summarize recent advances in immunotherapy strategies, focusing on Ebola targeting monoclonal antibodies (mAbs) that can be used for neutralization of virus and their potential use as passive prophylaxis.
In past few years, many epitopes on Ebola surface glycoproteins (GP) are identified. GP proteins play central role in virus entry inside host cells and pan out as effective target for mAb mediated neutralization. Various cocktails and mono-immunotherapy have been suggested with successful results in various laboratory organisms including rodents like mice and non-human primates (macaques). Studies postulated multiple potential targets for interference to inhibit viral entry. These sites are associated with conformation and proteolytic cleavage-based activation of GP protein and interaction with its endosomal receptor, Niemann Pick C1 (NPC1) (Saphire and Aman, 2016). Peripheral B-cells isolated from past Ebola virus outbreak human survivors followed by transformation of B-cells and screening Ebola binding antibodies, is one of the best methods for identification of immunologically safer anti-Ebola mAbs. One of the first mAb developed KZ52 was also used to characterize neutralizing epitope within EBOV GP, consisting of residues at the GP1-GP2 interface (Lee et al., 2008). Similarly, cocktail of 3mAbs with trade name ZMappTM is also shown to be promising in clearance of viremia (Murin et al., 2014). Recently Zhao et al., 2017 identified several cross-neutralizing epitopes suggesting that pan Ebola or broad neutralizing antibodies (bNAbs) and cross-protective vaccines might be developed. In 2020 Fan P. et al., also reported various human origin monoclonal antibodies showing neutralizing effect.
Filovirus
The family Filoviridae is subclassified into three genera i.e. Ebolavirus, Marburgvirus and Cuevavirus (Kuhn et al. 2014). The genus Ebolavirus consists of five species: Zaire Ebolavirus (now known as EBOV), Sudan Ebolavirus (SUDV), Bundibugyo Ebola¬virus (BDBV), Tai Forest Ebolavirus (TAFV) and Reston Ebolavirus (RESTV).
The Marburgvirus genus currently consists of a two closely related virus types, Marburg Marburgvirus (MARV) and Ravn virus (RAVV) both classified as single recognized species Marburg Marburgvirus I (Amarasinghe et al. 2017). The Cuevavirus genus includes a single species, Lloviucuevavirus, with one member, Lloviu virus. Although genome of Cuevaviruswas dem¬onstrated to be present in bats in northern Spain still virus is not isolated (Negredo et al. 2011).
Disease symptoms
Ebola Virus infection is followed by initial nonspecific symptoms, such as fever, severe fatigue, weakness, and headache, sometime accompanied with a maculopapular rash. In the later phase other symptoms including nausea, vomiting, diarrhea, abdominal pain, and other appears. In severe cases lethal unexplained hemorrhage starts. Symptoms may appear anywhere from 2 to 21 days after exposure to Ebola, but the average is 8 to 10 days (Brown et al. 2017; CDC 2020).
Epidemiology
Ebola virus is one of the world’s most feared diseases with mortality rates up to 25% to 90%.First discovered in 1976 with simultaneous outbreaks in Nzara, South Sudan, and Yambuka, Democratic Republic of the Congo (previously called Zaire), by 2013 there had been more than 1700 cases with a case fatality rate ranging from 25% to 90%. Of the 5 strains of EBV 4 strains of virus infect humans, with Zaire Ebolavirus as the most commonly found causative agent in outbreaks (CDC).
EVD is so fatal that until 2014 it was thought to be a localized disease with limited risk of converting to large outbreak. The 2013 to 2016 EVD outbreak in West Africa turned out otherwise and made scientific community to rethink the potential of EVD. During 2013-2016 reports of cases imported in different countries were also very high. For the first time in the history of Ebola virus outbreaks, world witness the deadly potential of Ebola in term of population infected and area involved. Modern world easy air, land, and water connectivity make the spread of disease at more easier and faster rate. Cases were imported into, some with localized onward transmission in Mali (8), Nigeria (20), the United States (2), the United Kingdom (1), Senegal (1), and Spain (1), along with the repatriation of a further 24 patients to the United States and Europe. World Health Organization (WHO) declared the conclusion of the Public Health Emergency of International Concern in March 2016,more than 28,000 people had been infected, with more than 11,00 deaths (CDC).
Host animals
Limited numbers of studies are available to understand the ecology of Filovirus Studies suggest that Filovirus are zoonotic. Although Filovirus natural reservoir (s) still not clear, but recently Marburgvirus and Ebolavirus were detected in fruit bats in Africa. Marburgvirus has also been isolated in several occasions from Rousettus bats in Uganda. To date, the only wild nonhuman primates found infected with Ebola viruses were African great apes: western lowland gorillas (Gorilla gorilla gorilla) and central chimpanzees (Pan troglodytes troglodytes) with EBOV and western chimpanzees (Pan troglodytes verus) with Taï Forest Ebolavirus (TAFV) (Leroy et al. 2004a, 2004b, 2005). Mortalilty is also reported by Reston Ebolavirus (RESTV) in long-tailed macaques (Macacafascicularis) held under captivity. (Gogarten et al. 2017)
Human Transmission
Lawrence et al. (2017) reviewed our current understanding of Ebola virus transmission in humans. EBOV has been detected as virus particle or its RNA in a range of bodily fluids including blood, stool, semen, breast milk, saliva, sweat and tears. Transmission of disease from infected to healthy individual is believed to be via contact of these body fluids, fomites, droplets and aerosols. Experimenal models using NHPs have shown that EBOV is both highly infectious and contagious; it can use many administration routes including oral, conjunctival, submucosal and respiratory routes. EBOV have been detected in semen of survivors at least up to around 500 days (Diallo et al. 2016).
Viral genome and proteins
Ebolavirion contains non segmented around 19kb of linear, negative-sense, single-stranded RNA. Capping and tailing of RNA absent, although 3’ and 5’ UTRs are present. The viral genome encodes a message for 7 structural proteins and 1 non-structural protein in gene order of 3′ –leader (UTR)– NP – VP35 – VP40 – GP/sGP – VP30 – VP24 – L – trailer (UTR) – 5′ (figure 1). UTR regions contain sequences responsible for gene regulation and new virus assembly
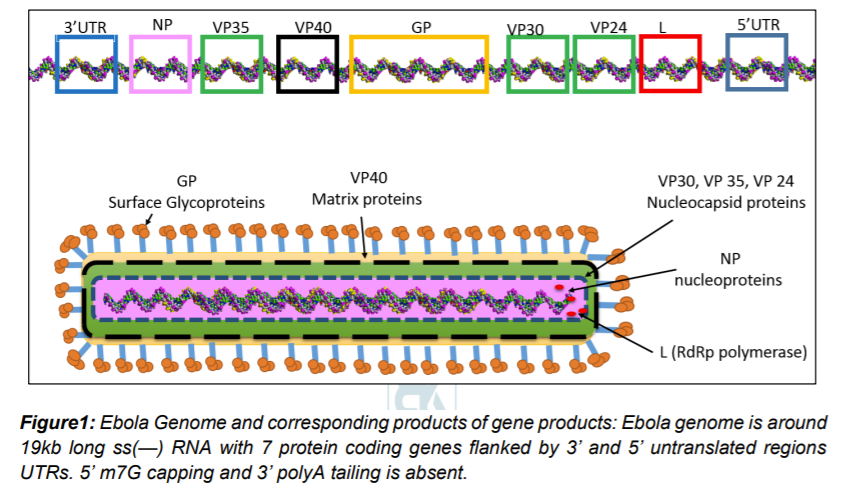
Understanding of Ebola Virus pathogenesis is used as a hallmark of Filovirus pathogenesis. Most of the information comes from experiments involving non-human primates. The disease severity in humans and NHPs appears to be an outcome of aggressive virus replication and inflammatory response by the host defense system. Possibly, various cytokines and cytotoxic molecules are released during Filovirus infection that results in disease symptoms, including high fever, vascular leakage and coagulopathy (Messaoudi et al. 2015)
Ebola genomic RNA is packed in nucleocapsid consisting of multiple proteins: NP (nucleoprotein), L (‘large’, polymerase), and viral proteins VP35, VP30, and VP24. Host derived membrane envelope has two transmembrane proteins i.e glycoprotein (GP) spike trimers and VP40 matrix protein (Beniac et al. 2017). GP trimers is a class I fusion protein and play role in attachment to host cells, endosomal entry, and membrane fusion. Many researchers have discovered antibodies targeting GP proteins and convincingly demonstrated the importance of GP protein as target for immunotherapy (Beniac et al. 2017).
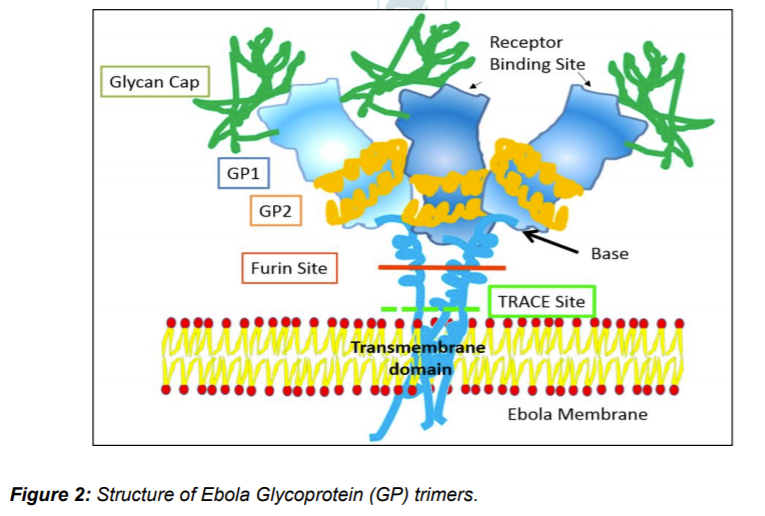
Experiments with GP expression in Ebola infected Vero E6 cell reveals that after transcription of GP gene it produces three type of GP proteins using post transcription RNA editing. Unedited mRNA (71%)istranslated to 364-residue soluble GP protein that later forms disulfide bridged dimers and a smaller peptide fragment called Δ (Delta) resulted due to furin cleavage of larger precursor of soluble GP. Around 24% GP mRNA are edited causing +1 shift in reading frame. The resulting +1 ORF translated to 676-residue full length GP that is further cleaved by Furin, forming membrane bound GP1/GP2 trimers. GP1 consists of RBS while GP2 wrap GP1 like a ribbon. The trimer structure forms chalice like structure and consists of glycan cap, receptor binding site (RBS), IFL cathepsin loop, Furin cleavage site, transmembrane domain, mucin like domain (M), TNF-α converting enzyme (TACE) cleavage site. Another 5% with +2 frameshift translated to much smaller soluble GP (ssGP). Both soluble GPs are N-linked glycosylated and present in easily detectable quantities body fluids. Δ peptide is O- linked glycosylated while GP1/GP2 trimers are both N- and O- linked glycosylated (Ning et al. 2016; Beniac et al. 2017). GP1/GP2 trimers are further cleaved to soluble GP trimers (GPCL), particles seen as GP shedding.
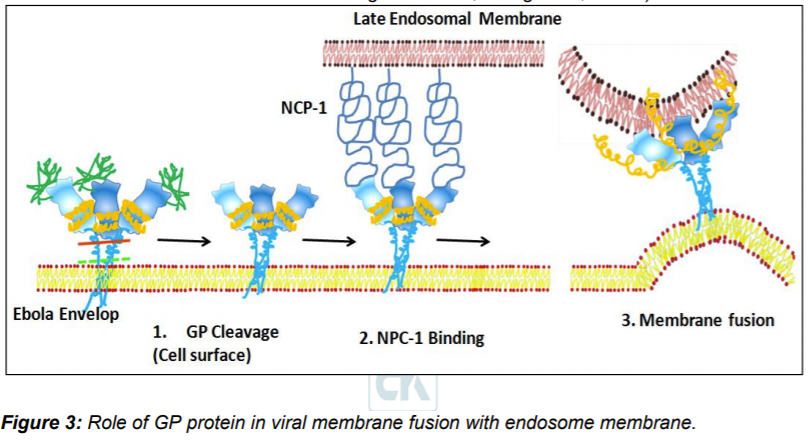
Virus particles attach to host cell through two types of relatively non-specific receptors: 1. Carbohydrate interacting C-type lectins (CLECs) and 2. Phosphatidylserine (PtdSer) receptors that interact with the viral envelope Phosphatidylserine. Other receptors like CLECs (LSECTin, DC-SIGN [dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin], L-SIGN [liver/lymph node-specific ICAM-3 grabbing nonintegrin], MBL mannose-binding lectin, and hMGL [human macrophage galactose- and N-acetylgalactosamine-specific C-type lectin]) are also thought to play role in viral glycans attachment. Micropinocytosis is thought to be primary entry route of Ebola virus. Initially virus particles are trapped in endosomes followed by environmental changes in form of lowering of pH. Lower pH and possibly activation of some hydrolytic enzymes triggers conformation changes in GP1/GP2 trimers leading to exposure of receptor binding domains on GP1.
In endosome Nieman-Pick disease type C1 (NPC-1)- protein is demonstrated as receptors for GP. Interaction with NPC-1 promotes further conformation changes in GP2 leading to exposure of fusion loop that embedded in endosomal membrane leading to fusion of viral and endosomal membrane. The viral genomics released in cytosol to proceed next stages of virion replication cycles leading to generation of new virus particle (Bornholdt et al., 2016a; Miller et al., 2012; Wang et al., 2016a).
Diagnostic methods
Initial phase of Ebola infection causes generalized symptoms like fever, making it difficult to suspect Ebola symptomatically. Suspected patients can me diagnosed by any of the available methods as prescribed by CDC, USA. The test includes: 1. for within a few days after symptoms begin: Antigen-capture enzyme-linked immunosorbent assay (ELISA) testing, IgM ELISA, Polymerase chain reaction (PCR) and Virus isolation; 2. Later in disease course or after recovery: IgM and IgG antibodies and 3. Retrospectively in deceased patients: Immunohistochemistry testing, PCR and Virus isolation (CDC, USA).
Immunotherapy
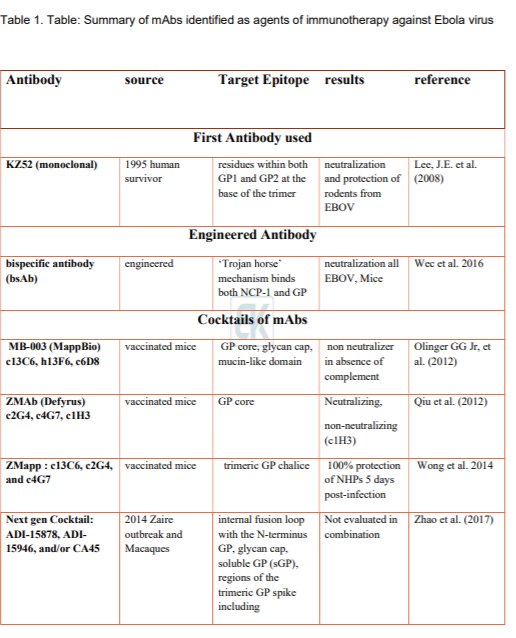
Importance of GP in virus entry make it most important target for the development of immunotherapy. Currently, three modes of viral targets are identified: 1. Inhibition of receptor binding, 2. Cathepsin-mediated cleavage inhibition, and 3. Blocking structural rearrangements of GP2 involved in formation of fusion loop. Monoclonal antibodies are currently most useful immunotherapy agents (Saphire and Aman 2017). In the past few years, many different antibodies were generated using EBOV specific B-cells isolated from human survivors. Other approaches for generation of mAbs using naïve antibody repertoire or somatic mutations were also successfully utilized. Summary of some antibodies are given in table 1. Similarly, epitopes discovered on GP1- GP2 trimer is given in Figure 4. One of the first antibodies developed was KZ52 that interacts with GP in its trimeric, pre-fusion conformation (GP1+GP2). The Ab was derived from B- cells of a human survivor of the 1995 Kikwit outbreak. KZ52 specific epitope were mapped to base of GP1/GP2 trime – the assembly responsible for fusion of viral membrane to endosome. KZ52 proved to be neutralizing ab in rodents but failed to protect EBOV-infected non-human primates (NHPs) (Lee et al. 2008).
Soon after the initial reports on anti EBOV antibody, idea of using cocktails of different antibodies were materialized. One of the first successful cocktails was developed by trade name ZMapp™, it consists of three monoclonal antibodies (mAbs). The mAbs 2G4 and 4G7, binds to epitope present at base of GP1/GP2 trimer, while mAb3C6 binds carbohydrate cap. ZMapp™ provides 100% protection of NHPs 5 days post-infection. ZMapp™ is the first successful candidate immunotherapy suggesting importance of combination of different target binding Ab, a key possible treatment for patients infected with EBV. This also shows importance of GP1/GP2 in viral entry and their importance as target for generation of neutralizing antibody (Wong et al. 2014).
Various other antibodies were also developed that specifically binds to epitopes present on different part of GP1/GP2 trimers. Here we tried to summarize some of these antibodies with information on their targets in very brief.
Furuyama et al. (2016) reported neutralizing mAb (6D6) that targets internal fusion loop in GP molecule thereby hindering membrane fusion of the viral envelope with endosomal membranes. The mAb recognize Ebolavirus glycoprotein, it effectively inhibits cellular entry all known Ebolavirus species in vitro. The mAb was successful in mouse models.
Flyak et al. (2016) isolated several antibodies specific to glycan cap that neutralized multiple Ebolaviruses, including SUDV as demonstrated in guinea pigs. The ab isolate from transformed B- cell isolated from human survivors of 2007 Uganda BDBV outbreak.
Bornholdt et al. (2016b) isolated and characterized 349 GP-specific monoclonal antibodies (mAbs). Antibodies were prepared from in vitro Epstein Barr virus mediated transformed B cells isolated from survived 2014 EBOV Zaire outbreak. They show 77% of the mAbs neutralize live EBOV, and several mAbs exhibit unprecedented potency. GP1/GP2 trimer stalk regions were appear to be the primary target for antibodies leading to inhibition of membrane fusion. The successful results were seen in mice.
Keck et al. (2016) identifies a set of pan-Ebolavirus and pan-Filovirus monoclonal antibodies (FVM02) derived from cynomolgus macaques. The macaques were challenged with mixture of GP and virus-like particles representing three different Filovirus species. Many different antibodies were isolated with different epitopes on GP. Antibody binding to a highly conserved epitope within the fusion loop of Ebolavirus and Marburgvirus species was also identified. Significant success in neutralizing EBOV was reported in mouse model of EBOV infection.
Everardo González-González (2020) describe the development of HEK293T cells engineered for stable expression of mAb13C6, a neutralizing anti-EBOV monoclonal antibody. The produced antibodies exhibited the expected functionality; they recognized the GP glycoprotein of the Ebola virus in both ELISA assays and cell binding experiments using HEK293T cells engineered to express the EBOV GP at their membrane surface.
Xiaoyan Tian (2020) reported the induction and isolation of two monoclonal antibodies that specifically recognized the glycoprotein (GP) and secreted glycoprotein (sGP) of the Ebola virus. Plasmids encoding either GP or sGP were constructed and immunized BALB/c mice, accordingly purified sGP was boosted. The antisera were analyzed for binding activity against sGP protein in enzyme‐linked immunosorbent assay (ELISA) and neutralization activity in a pseudo typed virus neutralization assay.
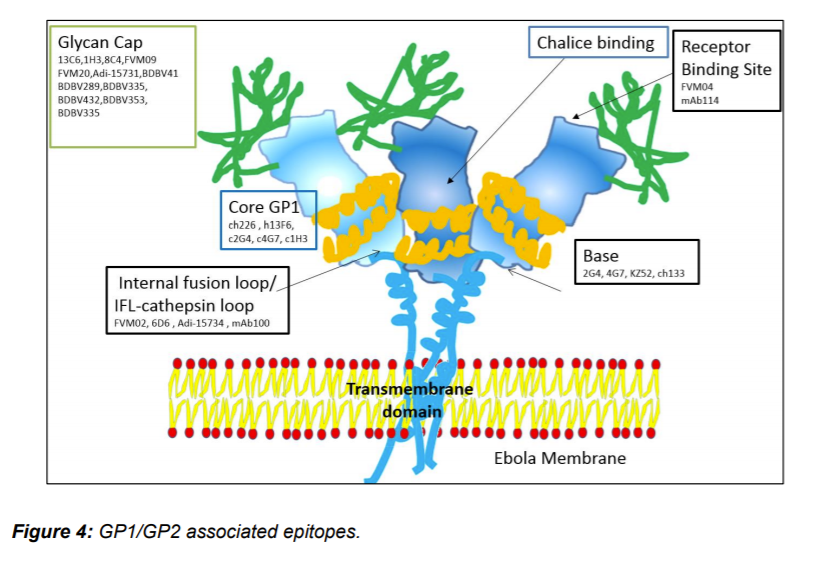
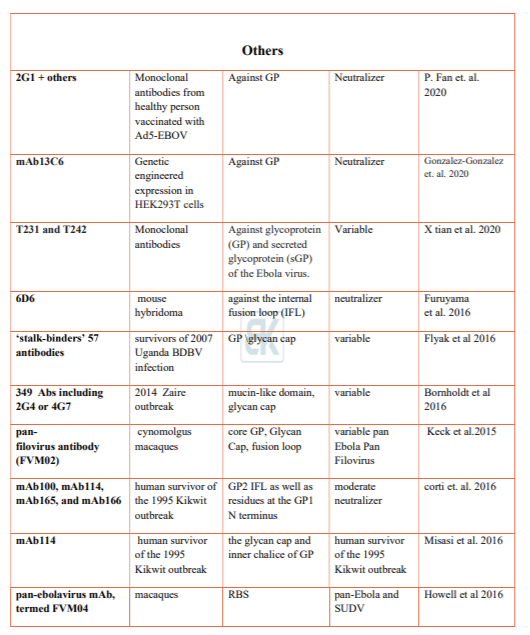
Corti et al. (2016) isolated peripheral B-cells from a survivor of the 1995 Kikwit, Democratic Republic of the Congo outbreak. One antibody (mAb100) provides protection in macaques up to 5 days post-infection of EBOV. Misasi et al. using crystal structures of antibodies bound GP reveals epitope near the tip of the GP2 IFL as well as residues at the GP1 N-terminus, which mediates viral cell entry. Interestingly mAb114 was shown interacting with the glycan cap and inner chalice of GP trimer. Even after removal of carbohydrate cap mAb114 remains associated. This binding may cause hindrance in interaction between GP RBS and NPC-1 leading to failure of membrane fusion.
Another pan Ebolavirus mAb, (FVM04) isolated from vaccinated macaque, identified by Howell et al. (2016), binds to the tip of the RBS crest and blocks NPC-1 binding .
Wec et al. (2017) screened their previously published 349 mAb library for broad neutralizers. Two mAbs were identified that could potently neutralize all five Ebolaviruses. The antibody targets the glycan cap, soluble GP (sGP), regions of the trimeric GP spike including the base, internal fusion loop (IFL) and the stalk. Prominent mAbs identified are the base and internal fusion loop (IFL) binders ADI-15878, ADI-15946, and ADI-15742, of these ADI-15878 and ADI-15742 shown highly potent neutralizing activity against all five known Ebolaviruses. ADI-16061 is the stalk binder.
Zhao et al. (2017) characterizes antibody (CA45) isolated immunized macaque. CA45 targets the internal fusion loop with the N-terminus GP. It potently neutralizes Ebola, Sudan, Bundibugyo, and Reston viruses; therefore, it is a candidate immunotherapy agent that can be used alone or in combination with other antibodies. It provided full protection against all pathogenic Ebolaviruses in mice, guinea pigs, and ferrets. Zhao et al. suggested that next generation cocktails of mAbs can comprise ADI-15878, ADI- 15946, and/or CA45.
Interestingly Wec et al. (2016) describe the use of ‘Trojan horse’ strategy using bispecific antibody, in which mAbs specific for NPC1 or the GP receptor-binding site are coupled to a mAb against a conserved, surface-exposed GP epitope. In mice, bispecific antibodies neutralized all ebolaviruses types.
Conclusion
In summary, these studies demonstrated the importance of GP proteins in viral entry into host cell. Various neutralizing epitopes are discovered on GP1, GP2 and glycan cap. Many different mAbs are identified and suggested to being used as monotherapy or cocktail. Antibodies like KZ52, 2G4, 4G7, 13C6, 6D6, FVM02, mAb100, mAb114, FVM04, CA45 and others successfully demonstrate the neutralization potential of these mAbs. Shedding of GP in body fluids may provide additional problem in use of mAbs. As most of the identified mAbs recognize epitopes on parts of GP trimer that is subject to cleaved during viral entry (GP shedding), this will provide the competition to circulating antibody and may lower the avidity of neutralizing antibodies.
It is suggested that cocktail of mAbs has higher potential to neutralize Ebola virus as compared to use of single mAb type. It is understandable that use of multiple antibodies that recognizes different target, have better chance than single antibodies. We should also explore these antibodies for their role in controlling the symptoms of disease in infected animals. This information will provide us the true potential of mAbs as agent of neutralization and passive prophylaxis in Ebola infected patient. At the moment we require a robust reagent that not only neutralize the Ebola Virus but also help in controlling disease associated symptoms. Use of neutralizing antibodies as passive prophylactic can be of great use to reduce mortality rates during future outbreaks. New approach like ‘Trojan horse’ bispecific antibodies have potential as broad anti-filovirus immune therapeutics. We should never underestimate the potential of other relatively silent Filovirus as agent of future outbreaks. Development of pan Ebola or pan Filovirus mAbs will surely be a step-in preparation for future.
References
1. Amarasinghe, Gaya K., YīmíngBào, Christopher F. Basler, SinaBavari, Martin Beer, NicolásBejerman, Kim R. Blasdell et al. “Taxonomy of the order Mononegavirales: Update 2017.” Archives of Virology (2017): 1-12.
2. Baseler, Laura, Daniel S. Chertow, Karl M. Johnson, Heinz Feldmann, and David M. Morens. “The pathogenesis of Ebola virus disease.” Annual Review of Pathology: Mechanisms of Disease 12 (2017): 387-418.
3. Beniac, Daniel R., and Timothy F. Booth. “Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 Å resolution.” Scientific reports 7 (2017): 46374.
4. Bornholdt, Zachary A., Esther Ndungo, Marnie L. Fusco, Shridhar Bale, Andrew I. Flyak, James E. Crowe, KartikChandran, and Erica Ollmann Saphire. “Host-primed Ebola virus GP exposes a hydrophobic NPC1 receptor-binding pocket, revealing a target for broadly neutralizing antibodies.” MBio 7, no. 1 (2016a): e02154-15.
5. Bornholdt, Zachary A., Hannah L. Turner, Charles D. Murin, Wen Li, Devin Sok, Colby A. Souders, Ashley E. Piper et al. “Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak.” Science 351, no. 6277 (2016b): 1078-1083.
6. Brown, Colin S., Stephen Mepham, and Robert J. Shorten. “Ebola Virus Disease.” Clinics in Laboratory Medicine 37, no. 2 (2017): 269-284.
7. CDC: Centers for Disease Control and Prevention. Outbreaks chronology: Ebola hemorrhagic fever [Internet]. Atlanta (GA): CDC; 2014. Available at: http://www.cdc. gov/vhf/ebola/resources/outbreak-table.html. Accessed September 2, 2020.
8. Corti, Davide, John Misasi, SabueMulangu, Daphne A. Stanley, Masaru Kanekiyo, Suzanne Wollen, AuréliePloquin et al. “Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody.” Science 351, no. 6279 (2016): 1339-1342.
9. Diallo, Boubacar, DaoudaSissoko, Nicholas J. Loman, HadjaAïssatou Bah, Hawa Bah, Mary Claire Worrell, RamataSacko et al. “Resurgence of Ebola virus disease in Guinea linked to a survivor with virus persistence in seminal fluid for more than 500 days.” Clinical infectious diseases 63, no. 10 (2016): 1353-1356.
10. Fan, Pengfei, et al. “Potent neutralizing monoclonal antibodies against Ebola virus isolated from vaccinated donors.” Mabs. Vol. 12. No. 1. Taylor & Francis, 2020.
11. Flyak, A. I., X. Shen, C. D. Murin, H. L. Turner, J. A. David, M. L. Fusco, R. Lampley et al. “Cross-reactive and potent neutralizing antibody responses in human survivors of natural Ebolavirus infection. Cell164: 392–405.” (2016).
12. Furuyama, W., A. Marzi, A. Nanbo, E. Haddock, J. Maruyama, H. Miyamoto, M. Igarashi et al. “Discovery of an antibody for pan-ebolavirus therapy. Sci Rep 6: 20514.” (2016).
13. Gogarten, Jan F., Sebastien Calvignac‐Spencer, and Fabian H. Leendertz. “Ebola Virus Disease.” The International Encyclopedia of Primatology (2017).
14. Gonzalez-Gonzalez E, Palestino-Diaz I, Lopez-Pacheco F, Marquez-Ipiña AR, Lara-Mayorga IM, Trujillo-de Santiago G, Alvarez MM. Rapid and cost-effective development of stable clones for the production of anti-Ebola monoclonal antibodies in HEK293T cells. bioRxiv. 2020 Jan 1.
15. Howell, Katie A., XiangguoQiu, Jennifer M. Brannan, Christopher Bryan, Edgar Davidson, Frederick W. Holtsberg, Anna Z. Wec et al. “Antibody treatment of Ebola and Sudan virus infection via a uniquely exposed epitope within the glycoprotein receptor-binding site.” Cell reports 15, no. 7 (2016): 1514-1526.
16. Keck, Zhen-Yong, Sven G. Enterlein, Katie A. Howell, Hong Vu, Sergey Shulenin, Kelly L. Warfield, Jeffrey W. Froude et al. “Macaque monoclonal antibodies targeting novel conserved epitopes within filovirus glycoprotein.” Journal of virology 90, no. 1 (2016): 279-291.
17. Kuhn, Jens H., Kristian G. Andersen, Sylvain Baize, YīmíngBào, SinaBavari, Nicolas Berthet, Olga Blinkova et al. “Nomenclature-and database-compatible names for the two Ebola virus variants that emerged in Guinea and the Democratic Republic of the Congo in 2014.” Viruses 6, no. 11 (2014): 4760-4799.
18. Kuhn, Jens H., YīmíngBào, SinaBavari, Stephan Becker, Steven Bradfute, Kristina Brauburger, J. Rodney Brister et al. “Virus nomenclature below the species level: a standardized nomenclature for filovirus strains and variants rescued from cDNA.” Archives of virology 159, no. 5 (2014): 1229-1237.
19. Lawrence, Philip, Nicolas Danet, Olivier Reynard, Valentina Volchkova, and Viktor Volchkov. “Human transmission of Ebola virus.” Current opinion in virology 22 (2017): 51-58.
20. Lee, Jeffrey E., Marnie L. Fusco, Ann J. Hessell, Wendelien B. Oswald, Dennis R. Burton, and Erica Ollmann Saphire. “Structure of the Ebola virus glycoprotein bound to a human survivor antibody.” Nature 454, no. 7201 (2008): 177.
21. Leroy, E. M., P. Telfer, B. Kumulungui, P. Yaba, P. Rouquet, P. Roques, J-P. Gonzalez, T. G. Ksiazek, P. E. Rollin, and E. Nerrienet. “A serological survey of Ebola virus infection in central African nonhuman primates.” The Journal of infectious diseases190, no. 11 (2004): 1895-1899.
22. Leroy, Eric M., Brice Kumulungui, Xavier Pourrut, Pierre Rouquet, Alexandre Hassanin, Philippe Yaba, André Délicat, Janusz T. Paweska, Jean-Paul Gonzalez, and Robert Swanepoel. “Fruit bats as reservoirs of Ebola virus.” Nature 438, no. 7068 (2005): 575-576.
23. Leroy, Eric M., Pierre Rouquet, Pierre Formenty, Sandrine Souquière, Annelisa Kilbourne, Jean-Marc Froment, Magdalena Bermejo et al. “Multiple Ebola virus transmission events and rapid decline of central African wildlife.” Science 303, no. 5656 (2004): 387-390.
24. Messaoudi, Ilhem, Gaya K. Amarasinghe, and Christopher F. Basler. “Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus.” Nature reviews. Microbiology 13, no. 11 (2015): 663.
25. Miller, Emily Happy, GregorObernosterer, MatthijsRaaben, Andrew S. Herbert, Maika S. Deffieu, Anuja Krishnan, Esther Ndungo et al. “Ebola virus entry requires the host‐programmed recognition of an intracellular receptor.” The EMBO journal 31, no. 8 (2012): 1947-1960.
26. Misasi, John, Morgan SA Gilman, Masaru Kanekiyo, Miao Gui, Alberto Cagigi, SabueMulangu, DavideCorti et al. “Structural and molecular basis for Ebola virus neutralization by protective human antibodies.” Science 351, no. 6279 (2016): 1343-1346.
27. Murin, Charles D., Marnie L. Fusco, Zachary A. Bornholdt, XiangguoQiu, Gene G. Olinger, Larry Zeitlin, Gary P. Kobinger, Andrew B. Ward, and Erica Ollmann Saphire. “Structures of protective antibodies reveal sites of vulnerability on Ebola virus.” Proceedings of the National Academy of Sciences 111, no. 48 (2014): 17182-17187.
28. Negredo, Ana, Gustavo Palacios, Sonia Vázquez-Morón, Félix González, HernánDopazo, Francisca Molero, Javier Juste et al. “Discovery of an ebolavirus-like filovirus in europe.” PLoS pathogens 7, no. 10 (2011): e1002304.
29. Ning, Yun-Jia, Fei Deng, Zhihong Hu, and Hualin Wang. “The roles of ebolavirus glycoproteins in viral pathogenesis.” VirologicaSinica (2016): 1-13.
30. Olinger, Gene Garrard, James Pettitt, Do Kim, Cara Working, OgnianBohorov, Barry Bratcher, Ernie Hiatt et al. “Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques.” Proceedings of the National Academy of Sciences 109, no. 44 (2012): 18030-18035.
31. Qiu, Xiangguo, Jonathan Audet, Gary Wong, Stephane Pillet, Alexander Bello, Teresa Cabral, Jim E. Strong et al. “Successful treatment of Ebola virus–infected cynomolgus macaques with monoclonal antibodies.” Science translational medicine 4, no. 138 (2012): 138ra81-138ra81.
32. Saphire, Erica Ollmann, and M. JavadAman. “Feverish Quest for Ebola Immunotherapy: Straight or Cocktail?.” Trends in microbiology 24, no. 9 (2016): 684-686.
33. Tian X, Chen D, Wang H, Xu S, Zhu L, Wu X, Wu Z. The induction and characterization of monoclonal antibodies specific to GP of Ebola virus. Journal of medical virology. 2020 Aug;92(8):996-1006.
34. Wang, Han, Yi Shi, Jian Song, Jianxun Qi, Guangwen Lu, Jinghua Yan, and George F. Gao. “Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1.” Cell 164, no. 1 (2016): 258-268.
35. Wec, Anna Z., Andrew S. Herbert, Charles D. Murin, Elisabeth K. Nyakatura, Dafna M. Abelson, J. Maximilian Fels, Shihua He et al. “Antibodies from a Human Survivor Define Sites of Vulnerability for Broad Protection against Ebolaviruses.” Cell 169, no. 5 (2017): 878-890.
36. Wec, Anna Z., Elisabeth K. Nyakatura, Andrew S. Herbert, Katie A. Howell, Frederick W. Holtsberg, Russell R. Bakken, Eva Mittler et al. “A “Trojan horse” bispecific-antibody strategy for broad protection against ebolaviruses.” Science 354, no. 6310 (2016): 350-354.
37. Wong, Gary, XiangguoQiu, Gene G. Olinger, and Gary P. Kobinger. “Post-exposure therapy of filovirus infections.” Trends in microbiology 22, no. 8 (2014): 456-463.
38. Zhao, Xuelian, Katie A. Howell, Shihua He, Jennifer M. Brannan, Anna Z. Wec, Edgar Davidson, Hannah L. Turner et al. “Immunization-elicited broadly protective antibody reveals ebolavirus fusion loop as a site of vulnerability.” Cell 169, no. 5 (2017): 891-904.