Recent Advances in Rapid Diagnosis and Analysis of Infectious Agents in Human Blood Stream
Pradeep Kumar Singh1*, Shweta Kusmakar Singh2
1Department of Microbiology, S.M.M.H. Medical College, Saharanpur, UP, India
2Department of Medical Oncology, All India Institute of Medical Sciences, New Delhi, India
Abstract
The ability of rapid diagnosis and analysis of infectious agents such as pathogens in human blood stream is considered pivotal and an important step for patient treatment and effective monitoring. This is also essential to ensure the safety of the donor blood supply. This has driven research in medical biotechnology and the focus has been on developing rapid diagnostic ability that enables detect and identify pathogens in blood borne infections. This research seeks to mitigate the challenges involving infections such as pathogens induced sepsis. These infections are believed to be a major cause of mortality in hospitalized patients worldwide. It is of general consensus that establishing a rapid diagnosis would not only enable an early and adequate antimicrobial therapy but it would also result in positive clinical outcomes for patients. In this perspective, we have presented a brief overview of some of the notable recent advances in diagnostics to detect infectious agents in human blood.
Keywords: Diagnostics, infectious diseases, pathogens, viruses, lasers, blood borne pathogens, spectrometry
DOI: https://doi.org/10.37756/bk.22.4.2.1
Article type: Perspective
Received: December 15, 2021
Revised: January 10, 2022
Accepted: January 15, 2022
Please cite this article as: Singh PK, Recent Advances in Rapid Diagnosis and Analysis of Infectious Agents in Human Blood Stream, Biotechnol. kiosk, Vol 4, Issue 2, PP: 1-11 (2022); DOI: https://doi.org/10.37756/bk.22.4.2.1
*Corresponding author: Dr. Pradeep Kumar Singh
E-mail address: pksingh1976a@gmail.com
Introduction
In battling against complex diseases, a significant public and medical concern is the quality and safety of blood products that are used in transfusions. It is believed that blood transfusion-transmitted infections or infectious agents can result in serious medical conditions such as sepsis. This can lead to fatal consequences especially in highly vulnerable populations including neonates, the elderly, or immunocompromised persons. Studies conducted over the last decades have shown emerging pathogens that can severely affect blood safety. To battle against such infectious pathogens, the focus has been to employ rapid molecular tests for detection of blood-borne pathogens such as West Nile virus, dengue virus (DENV), and Babesia microti etc. [1, 2].
There are several existing processes of blood culture that are employed for the detection of an infection and the identification of the responsible organism for diagnosis of human blood stream infections. However, most of these processes involve a fairly long time period to complete the tests. Hence, it is not rapid at times that usually influences vital treatment decisions. The time-consuming process of blood culture is due to the fact that it involves steps starting from micro-organisms that are present in a blood sample and are enriched in cultivation medium in blood culture bottles under continuous monitoring. In the next step, Gram staining is done that allows the first adjustment of antibiotic therapy. Additionally, sub cultivation may also be required for identification and antimicrobial susceptibility testing. This entire process of blood culture therefore becomes quite complex that could essentially involve up to total 72 hours depending on the pathogen. This also includes the use of highly skilled personnel and complex sample preparation. Furthermore, the process becomes even more complex and time consuming in the event of transporting the blood sample to a microbiology laboratory for advanced analysis. All these complexities have triggered research in medical biotechnology to develop next generation diagnostics that ensures prevention of fatal diseases that are caused by pathogens in blood born infections [3, 4].
To this end, medical technologists and biotechnologists have been involved in cutting edge research to develop diagnostic technology mostly based on lasers and spectrometry that gives the ability to rapidly (within minutes) diagnose blood borne infections on-site. It is believed that on-site diagnostic technology is transformative as it requires simple sample preparation and no requirement for highly skilled personnel. This significantly enhances the ability to identify, contain, and treat blood borne infections. This is expected to greatly reduce the time needed to screen donated blood for infections. Researchers envision that the development of laser-based blood diagnostic technology with these capabilities could pave the way to future breakthroughs that would take the laboratory diagnosis to the next level, where patients could be screened for infections and receive prescribed treatments immediately [5-9].
Here, we have described some of the recent significant advances in rapid diagnostic technologies for quick analysis of human blood.
1. Ability to Detect Parasites, Bacteria and Viruses in Human Blood: Laser-Based Point-of-Care Diagnostic Technology
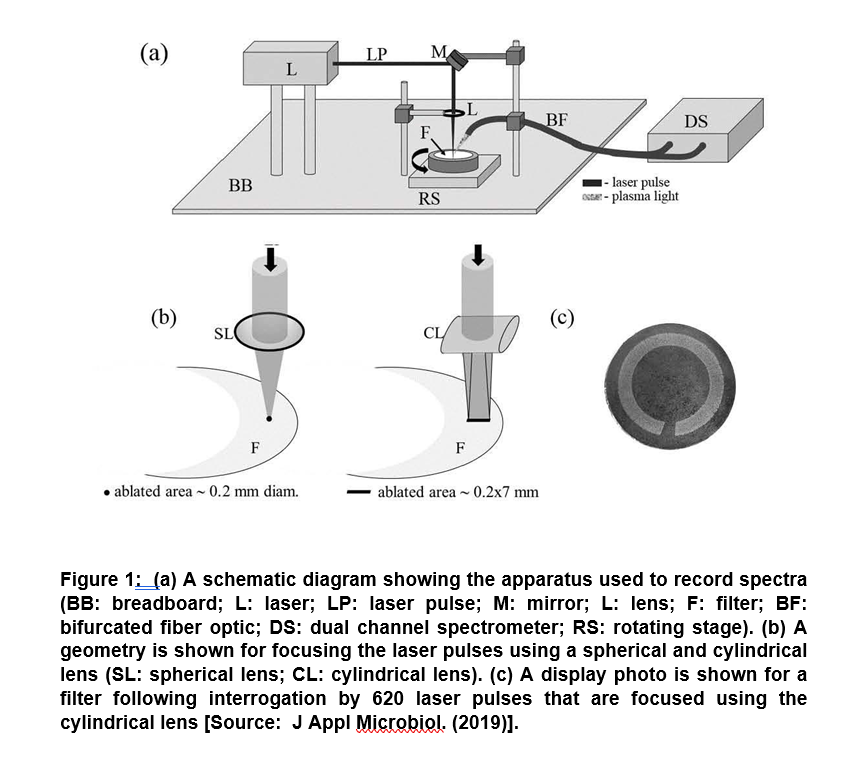
In a recent study of major significance, researchers showed the potential of a laser-based diagnostic method to detect the presence of parasites, bacteria and viruses in human blood. The technique was based on Laser-Induced Breakdown Spectroscopy (LIBS). It showed the capability of point-of-care diagnostic based on the analysis of spectra collected from a series of laser sparks that were formed on a blood sample. In this method, a laser pulse is used to simultaneously vaporize a small sample mass that subsequently excites the resulting atoms. This results in emitting light formation of a hot plasma on the sample surface. Finally, light from the plasma is collected and spectrally resolved and subsequently recorded for spectra (Figure 1) [8].
The technique was shown to be capable of generating analysis results within minutes. Researchers showed the presence of pathogens in the blood using a novel analysis approach and successfully demonstrated to clinically relevant levels of 10 cells, copies, or parasites per ml. It showed the differentiation of blood spiked with viruses, bacteria, or protozoan parasites to clinically relevant levels in six blood types (O+, O, AB+, A+, A-, B+). These results pave the way to simultaneously detect multiple pathogens in blood that could be employed for more rapid blood analysis in the future [8].
2. Rapid Diagnostic Technology for Human Visceral Leishmaniasis (Kala-Azar)
Leishmaniasis is considered a deadly disease. This disease is believed to be endemic in 98 countries, with the highest burden of disease in India, Brazil and other developing nations. Studies on Leishmaniasis have suggested that the disease is caused by protozoan parasites of the genus Leishmania and the most severe form of this disease is Visceral Leishmaniasis (VL), which is also known as Kala-Azar. It has been suggested that Kala-Azar is caused by L. chagasi/L. infantum in the Americas and L. donovani and L. infantum in Afro-Eurasia. Rapid and precise diagnosis of human VL is the suggested treatment pathway to battle against human VL, which is otherwise believed to be a fatal disease. To this end, researchers believe that for VL eradication, it is essential to develop novel, effective and affordable assays for rapid diagnosis of VL [10-13].
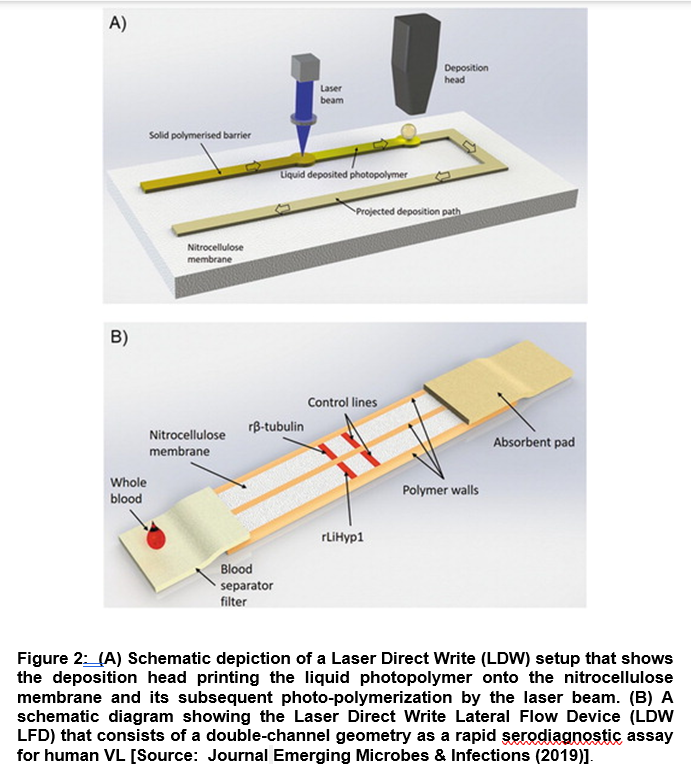
In a recent study, researchers demonstrated a novel diagnostic technique based on Laser Direct-Write (LDW) technology coupled with a Lateral Flow Device (LFD) with double-channel geometry. The advantage of this diagnostic technology lies in the fact that it can work on a low-cost paper platform that can be leveraged for a rapid and accurate serodiagnostic assay for human VL. Researchers employed the Duplex VL-LFD that was based on a laser-patterned microfluidic device using two recombinant Leishmania proteins, β-tubulin and LiHyp1, as novel diagnostic antigens (Figure 2) [14].
They further tested the VL-LFD assay with blood/serum samples from patients diagnosed with VL, Tegumentary Leishmaniasis and Leishmaniasis of unknown identity. The study also involved other parasitic diseases with similar clinical symptoms. Using LBW-LFD technique, out of the 22 positive samples from patients that were diagnosed clinically with VL infection, 20 tested positive with the Duplex VL-LFD, compared to 17 patients that were tested positive with the available commercial kit [14]. Further, researchers showed the better performance of LBW-LFD technique compared to the standard methods to diagnose and screen patients with VL. It is envisioned that the applications of this new VL-LFD diagnostic technique could be extended to employ to other comparative testing in larger patient groups focusing in areas with endemic VL. This could be a significant step forward to improve diagnosis and disease management of Kala-Azar [14].
3. Identification of Bacteria in Human Bloodstream by Immunoaffinity Mass Spectrometry
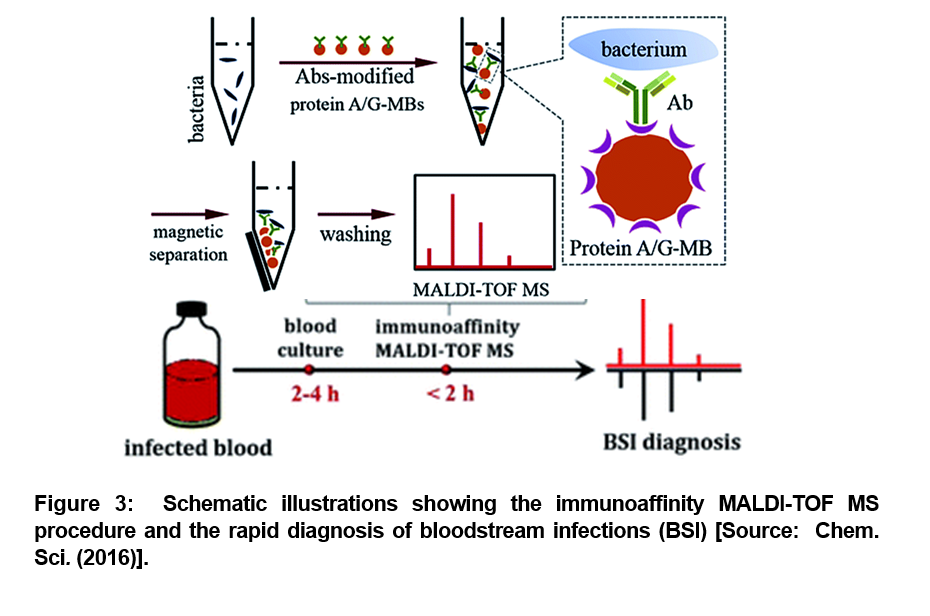
As we described in the beginning, blood stream infections (BSI) associated sepsis can lead to serious medical conditions. BSI is caused by the presence of bacteria or fungi in the blood stream. According to one estimate, about 600,000 BSI episodes occur in North America every year, and about 1, 200,000 BSI episodes affect Europe. This results in roughly 86,000 and 157,000 deaths, respectively. To mitigate the challenges posed by BSI, a rapid blood test and diagnosis is recomended to effectively battle such bacterial infections in the blood stream [15-18]. To this end, researchers recently demonstrated a sensitive method to rapidly and accurately identify bacteria in human blood samples. They employed a combined optimized matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and efficient immunoaffinity enrichment/separation method for the rapid diagnosis (Figure 3). A proof-of-concept level demonstration was reported with whole blood spiked with a low initial concentration (102 or 103 cells per mL) of bacteria that was cultured in commercial blood culture bottles and analyzed by the developed method after different blood culture times [19].
The employed method showed a distinct advantage of high sensitivity. It was shown that the blood culture time required for diagnosis could be significantly reduced. The bacteria was successfully identified after 4 hours of blood culture (Figure 3) [19]. This method was further optimized for the development of an entirely new diagnostic process that could be accurately accomplished within half a day from start of the tests to the completion of the process. This paves the way for employing immunoaffinity mass spectrometry method in all future studies that could facilitate anti-bacterial therapy in a timely fashion to mitigate the risk of mortality from bloodstream infections [19].
4. Case Study on Rapid Diagnosis of Bloodstream Infections in Critical Patients
Previous studies have shown that prompt treatment with targeted antibiotics can be leveraged to not only mitigate the challenges of the financial impact but also positively impact the clinical outcome of bloodstream infection and associated sepsis that represent a major source of mortality in industrialized countries [20-27]. Researchers conducted a case study to assess the usefulness of the IRIDICA BAC BSI Assay, a PCR/ESI-MS-based technology for the early diagnosis of bloodstream infections from primary blood samples in critical patients [28]. This evaluation was performed by comparison with the traditional culture-based methods. In this study, researchers investigated a total of 300 prospective whole blood specimens obtained from patients suspected of sepsis (Figure 4). They found that the overall concordance between the two techniques was of 86%, with a calculated sensitivity of 76% and an assay specificity of 90%. Further, the clinical significance of discrepant results was evaluated reviewing the patients’ clinical records and the results of additional relevant microbiological tests [28].

Figure 4: Distribution of the organisms is shown. The organisms reported by culture (solid bar) and PCR/ESI-MS (patterned bar) are sorted by decreasing order of PCR/ESI-MS reported organisms [Source: PLoS One (2018)]. This case study was significant as the data supported the ability of the IRIDICA BAC BSI Assay to identify a broad range of bacteria directly from primary whole blood samples, within eight hours, which implied a timely administration of a suitable treatment [28].
Concluding Remarks
In conclusion, we have described the requirements for new clinical diagnostics for rapid detection of pathogens in human blood. Such rapid identification has been shown to be useful in combating complex diseases that are caused by blood pathogens. This can also be leveraged to adapt to newly emerging infectious agents and to screen simultaneously for multiple infectious agents in bloodstream. To this end, several studies have shown promise of laser-assisted diagnostics and the unique capability of laser technology for rapid identification of infections in bloodstream. This new diagnostic technology platform is expected to transform the existing blood culture process to a far more superior and rapid process of identification of infections within a few hours and also on-site. The laser-based approaches can be leveraged to achieve sensitive, multiplex detection that requires minimal sample preparation while at the same time providing rapid results that are generated within minutes. Thus, this versatile point-of-care diagnostics offers a disruptive technology platform that could potentially be a game changer in next generation diagnostics and therapeutics industry. Recent studies have suggested that these application-relevant properties can be leveraged to boost the diagnostic abilities. This can be further equipped with the additional features such as flexibility in the technology that allows to add new agent detection by simply adjusting the detection programming. These steps are considered critically important to achieve the ultimate goal of superior clinical diagnosis of bloodstream infections along with prevention of associated medical fatalities in the future.
References
[1] Grigorenko E, Fisher C, Patel S, Winkelman V, Williamson P, Chancey C, Añez G, Rios M, Majam V, Kumar S and Duncan R, Highly Multiplex Real-Time PCR-Based Screening for Blood-Borne Pathogens on an OpenArray Platform. J Molecular Diagn 2017, 19:549–560, DOI: https://doi.org/10.1016/j.jmoldx.2017.03.004.
[2] Grigorenko E, Fisher C, Patel S, Chancey C, Rios M, Majam V, Nakhasi HL and Duncan R, Multiplex Screening for Blood-Borne Viral, Bacterial, and Protozoan Parasites using an OpenArray Platform. J Molecular Diagn 2014, 16:136–144, DOI: https://doi.org/10.1016/j.jmoldx.2013.08.002.
[3] Kourout M, Fisher C, Purkayastha A, Tibbetts C, Winkelman V, Williamson P, Nakhasi HL and Duncan R, Multiplex detection and identification of viral, bacterial, and protozoan pathogens in human blood and plasma using a high-density resequencing pathogen microarray platform. Transfusion 2016, 56:1537–1547, DOI:
https://doi.org/10.1111/trf.13524.
[4] Dong M, Fisher C, Anez G, Rios M, Nakhasi HL, Hobson JP, Beanan M, Hockman D, Grigorenko E and Duncan R, Standardized methods to generate mock (spiked) clinical specimens by spiking blood or plasma with cultured pathogens. J Applied Microbiol 2016, 120:1119–1129, DOI: https://doi.org/10.1111/jam.13082.
[5] Diedrich J, Rehse SJ and Palchaudhuri S, Escherichia coli identification and strain discrimination using nanosecond laser-induced breakdown spectroscopy. Appl Phys Lett 2007, 90:163901, DOI: https://doi.org/10.1063/1.2723659.
[6] Marcos-Martinez D, Ayala JA, Izquierdo-Hornillos RC, Manuel de Villena F.J. and Caceres JO, Identification and discrimination of bacterial strains by laser induced breakdown spectroscopy and neural networks. Talanta 2011, 84:730–737, DOI: https://doi.org/10.1016/j.talanta.2011.01.069.
[7] Multari RA, Cremers DA, Bostian ML, Dupre JM and Gustafson JE, Proof-of-Principle for a Real-Time Pathogen Isolation Media Diagnostic: The Use of Laser-Induced Breakdown Spectroscopy (LIBS) to Discriminate Bacterial Pathogens and Antimicrobial-Resistant Staphylococcus aureus Strains grown on Blood Agar. J Pathogens, 2013a, Article ID 898106, 10.1155/2013/898106, DOI: https://doi.org/10.1155/2013/898106.
[8] Multari RA, Cremers DA, Nelson A, Karimi Z, Young S, Fisher C, Duncan R, The use of laser-based diagnostics for the rapid identification of infectious agents in human blood. J Appl Microbiol. 2019, 126(5):1606-1617, DOI: https://doi.org/10.1111/jam.14222.
[9] Klein S, Zimmermann S, Ko¨hler C, Mischnik A, Alle W and Bode KA., Integration of matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry in blood culture diagnostics: a fast and effective approach. Journal of Medical Microbiology 2012, 61:323–331, DOI: https://doi.org/10.1099/jmm.0.035550-0.
[10] Chappuis F, Sundar S, Hailu A, et al., Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol 2007, 5(11):873–882, DOI: https://doi.org/10.1038/nrmicro1748.
[11] Elmahallawy EK, Sampedro Martinez A, Rodriguez-Granger J, et al., Diagnosis of leishmaniasis. J Infect Dev Ctries 2014, 8(8):961–972, DOI: https://doi.org/10.3855/jidc.4310.
[12] Cota GF, de Sousa MR, de Freitas Nogueira BM, et al., Comparison of parasitological, serological, and molecular tests for visceral leishmaniasis in HIV-infected patients: a cross-sectional delayed-type study. Am J Trop Med Hyg 2013, 89(3):570–577, DOI: https://doi.org/10.4269/ajtmh.13-0239.
[13] de Paiva-Cavalcanti M, de Morais RC, Pessoa ESR, et al., Leishmaniases diagnosis: an update on the use of immunological and molecular tools. Cell Biosci 2015, 5:31, DOI: https://doi.org/10.1186/s13578-015-0021-2.
[14] Humbert MV, Costa LE, Katis I, Fonseca Ramos F, Sanchéz Machado A, Sones C, Ferraz Coelho EA, Christodoulides M, A rapid diagnostic test for human Visceral Leishmaniasis using novel Leishmania antigens in a Laser Direct-Write Lateral Flow Device. Emerg Microbes Infect 2019, 8(1):1178-1185, DOI: https://doi.org/10.1080/22221751.2019.1635430.
[15] Peters RP, van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke-Grauls CM, New developments in the diagnosis of bloodstream infections. Lancet Infect Dis 2004, 4(12):751-60, DOI: https://doi.org/10.1016/S1473-3099(04)01205-8.
[16] Kang DK, Ali MM, Zhang K, Huang SS, Peterson E, Digman MA, Gratton E, Zhao W, Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nat Commun 2014, 5:5427, DOI: https://doi.org/10.1038/ncomms6427.
[17] Holland RD, Wilkes JG, Rafii F, Sutherland JB, Persons CC, Voorhees KJ, Lay JO Jr, Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 1996, 10(10):1227-32, DOI: https://doi.org/10.1002/(SICI)1097-0231(19960731)10:10<1227::AID-RCM659>3.0.CO;2-6.
[18] Croxatto A, Prod’hom G, Greub G, Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev 2012, 36(2):380-407, DOI: https://doi.org/10.1111/j.1574-6976.2011.00298.x.
[19] Zhu Y, Qiao L, Prudent M, Bondarenko A, Gasilova N, Möller SB, Lion N, Pick H, Gong T, Chen Z, Yang P, Lovey LT, Girault HH, Sensitive and fast identification of bacteria in blood samples by immunoaffinity mass spectrometry for quick BSI diagnosis. Chem Sci. 2016, 1;7(5):2987-2995, DOI: https://doi.org/10.1039/c5sc04919a.
[20] Riedel S, Carroll KC, Laboratory detection of sepsis: biomarkers and molecular approaches. Clin Lab Med 2013, 33(3):413–443, DOI: https:doi.org/10.1016/j.cll.2013.03.006.
[21] Lodes U, Bohmeier B, Lippert H, König B, Meyer F, PCR-based rapid sepsis diagnosis effectively guides clinical treatment in patients with new onset of SIRS. Langenbecks Arch Surg 2012, 397(3):447–455, DOI: https://doi.org/10.1007/s00423-011-0870-z.
[22] Metzgar D, Frinder MW, Rothman RE, Peterson S, Carroll KC, Zhang SX, et al., The IRIDICA BAC BSI Assay: Rapid, Sensitive and Culture-Independent Identification of Bacteria and Candida in Blood. PLoS One 2016, 11(7): e0158186, DOI: https://doi.org/10.1371/journal.pone.0158186.
[23] Lyle N, Boyd J, The potential for PCR based testing to improve diagnosis and treatment of sepsis. Curr Infect Dis Rep 2013, 15(5):372–379, DOI: https://doi.org/10.1007/s11908-013-0350-4.
[24] Peker N, Couto N, Sinha B, Rossen JW, Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: recent developments in molecular approaches. Clinical Microbiology and Infection 2018, 24 (9):944-955, DOI: https://doi.org/10.1016/j.cmi.2018.05.007.
[25] Perera RS, Ding XC, Tully F, Oliver J, Bright N, Bell D et al.,
Development and clinical performance of high throughput loop-mediated isothermal amplification for detection of malaria. PLoS One 2017,12:e0171126, DOI: https://doi.org/10.1371/journal.pone.0171126.
[26] Peri AM, Stewart A, Hume A et al., New Microbiological Techniques for the Diagnosis of Bacterial Infections and Sepsis in ICU Including Point of Care. Curr Infect Dis Rep 2021, 23:12, DOI: https://doi.org/10.1007/s11908-021-00755-0.
[27] Vincent JL, Brealey D, Libert N, Abidi NE, O’Dwyer M, Zacharowski K, Mikaszewska-Sokolewicz M, Schrenzel J, Simon F, Wilks M, Picard-Maureau M, Chalfin DB, Ecker DJ, Sampath R, Singer M, Rapid Diagnosis of Infections in the Critically Ill Team. Rapid Diagnosis of Infection in the Critically Ill, a Multicenter Study of Molecular Detection in Bloodstream Infections, Pneumonia, and Sterile Site Infections. Crit Care Med 2015, 43(11):2283-91, DOI: https://doi.org/10.1097/CCM.0000000000001249.
[28] Tassinari M, Zannoli S, Farabegoli P, Pedna MF, Pierro A, Mastroianni A, Fontan R, Luongo L, Sarnataro G, Menegatti E, Caruso A, Sambri V, Rapid diagnosis of bloodstream infections in the critically ill: Evaluation of the broad-range PCR/ESI-MS technology. PLoS One 2018, 13(5):e0197436, DOI: https://doi.org/10.1371/journal.pone.0197436.