
Abstract
The current COVID-19 pandemic has triggered innovations in therapeutic strategies that are thought to provide viable and faster pathways to cure infected people by the deadly virus, SARSCoV-2. In this review, we have described the known epidemiology along with the discovered genome structure and viral factors of SARS-CoV-2. Further, the latest innovations in therapeutics including significant breakthroughs in clinical trials on antiviral drugs, rendesivir and chloroquine including the combination drugs have been highlighted. This brief overview on therapeutic strategies should serve a medium to researchers for further innovations in drug discovery and repurposing of anti-viral drugs for SARS-CoV-2.
*Email: shyabiswas@biotechkiosk.com
SARS-CoV-2: Epidemiology, Genome Structure and Key Viral Factors
The severe acute respiratory syndrome coronavirus (SARS-CoV-2; COVID-19) epidemic broke out first in Wuhan, China in December 2019 that later became a worldwide deadly pandemic. Bat was suggested in earlier studies as the potential reservoir of SARS-CoV-2 [1]. Detailed virus genome sequencing of COVID-19 was conducted throughout the genome to Bat CoV RaTG13 that showed 96.2% overall genome sequence identity of SARS-CoV-2. The virus genome sequencing results and evolutionary analysis suggested that bat CoV and human SARS-CoV-2 might share the same ancestor. Thus, bat was a suspect as natural host of virus origin, and it was thought that SARS-CoV-2 got transmitted from bats via unknown intermediate hosts to infect humans [1, 2]. Further, researchers estimated the basic reproduction number (R0) of SARS-CoV-2 to be around 2.2, or even more (range from 1.4 to 6.5) in familial clusters of pneumonia outbreaks. This has led to a steady spread of COVID-19 across the globe by human-to-human transmissions taking tens of thousands of lives so far [3, 4].
Researchers have divided coronaviruses (CoV) into four genera that include α−/β−/γ−/δ-CoV. Among these different types of viruses, it has been shown that α- and β-CoV are able to infect mammals, while γ- and δ-CoV tend to infect birds [1]. The SARS-CoV-2 has been identified as a β-coronavirus, which is enveloped in a non-segmented positivesense RNA virus (Figure 1) [1]. In previous studies, researchers identified six CoVs as human-susceptible virus. In these six CoVs, the group of α-CoVs – HCoV-229E and HCoV-NL63, and the group of β-CoVs – HCoV-HKU1 and HCoV-OC43 were shown with low pathogenicity causing mild respiratory symptoms similar to a common cold [1]. However, the two other known βCoVs – SARS-CoV and MERS-CoV have been shown to cause severe and potentially fatal respiratory tract infections [1]. Some of the latest studies have clearly suggested involvement of the receptor angiotensinconverting enzyme 2 (ACE2) that is used by SARS-CoV-2 to infect humans (Figure 1) [1, 2].
Regarding genome structure and key viral factors, Figure 1 schematically illustrates the genome of CoV-2 virus that contains a variable number (6–11) of open reading frames (ORFs) [1]. Researchers have shown that two-thirds of viral RNA, that is mainly located in the first ORF (ORF1a/b) translates two polyproteins, pp1a and pp1ab, and encodes 16 non-structural proteins (NSP) with the remaining ORFs encode accessory and structural proteins (Figure 1) [1]. Further, the rest of the virus genome has been shown to encode four essential structural proteins, including spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein. It has also been shown that in addition to structural proteins, several accessory proteins can interfere with the host innate immune response [1].
Therapeutic Strategies for COVID-19: The Important Roles of Anti-Viral Drugs
Innovations in therapeutic strategies are thought to provide viable pathways to a faster solution to the current COVID-19 pandemic to cure infected people and also to prevent further epidemics.
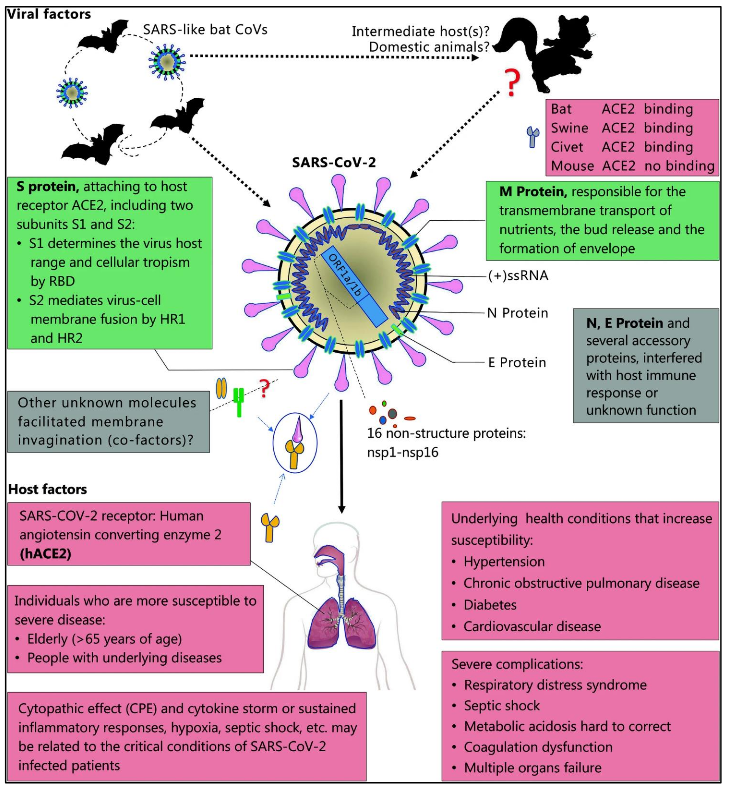
Figure 1: The pathogenesis of SARS-CoV-2Viral that is influenced by host factors. It shows illustration of bats that are considered the reservoir of SARS-CoV-2. The novel virus is thought to have originated from bats or unknown intermediate hosts and subsequently cross the species barrier into humans with virus-host interactions that affect viral entry and replication. The virus genome encodes spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein, and also several accessory proteins. Host factors are shown in the lower panel [Source: Military Med Res (2020)].
The promising strategies include therapeutic agents targeting nucleosides, nucleotides, viral nucleic acids and enzymes/proteins involved in the replication and transcription of SARS-CoV-2 [5]. To this end, repurposing some of the existing antiviral agents to treat infection from SARS-CoV-2 are already moving into clinical trials. Figure 2 shows potential drug targets for beta-coronaviruses [6].

Figure 2: Promising drug targets for beta-coronaviruses. (a) Genomic organization of SARSCoV-2 (the coding regions for proteins for potential drug targets). (b) A drug binding pocket along with chemical structures of potential inhibitors [Source: Nat Rev Drug Discov. (2020)].
Researchers have shown potentials of approved nucleoside analogues (favipiravir and ribavirin) and experimental nucleoside analogues (remdesivir and galidesivir) against SARS-CoV-2 (Figure 2) [6]. Previous studies have suggested that nucleoside analogues in the form of adenine or guanine derivatives can target the RNA-dependent RNA polymerase and block viral RNA synthesis in a broad spectrum of RNA viruses, including human coronaviruses [5, 6]. For example, researchers have shown Favipiravir (T-705), which is a guanine analogue approved for influenza treatment, can effectively inhibit the RNA-dependent RNA polymerase of a range of RNA viruses including influenza, Ebola, yellow fever, chikungunya, norovirus and enterovirus. In this regard, a recent research reported the promising activity of Favipiravir against COVID-19 (EC50 = 61.88 μM in Vero E6 cells) [1, 5-7].
Remdesivir (GS-5734, Figure 2), which is essentially a 1′-cyano-substituted adenosine nucleotide analog prodrug that inhibits viral RNA polymerases has shown quite a lot of promise. This anti-viral drug has been tested for a wide ranging antiviral activity against several RNA viruses [1]. In a most recent research, in-vitro activity of Remdesivir was demonstrated against SARS-CoV-2, and two-thirds of severe COVID-19 cases improved on administering remdesivir (Figure 3) [8]. In this study that employed a cohort of patients hospitalized for severe COVID-19 who were treated with compassionate-use remdesivir, clinical improvement was observed in 36 of 53 patients (68%) in the category of oxygen support, while Improvement was observed in all 12 patients who were breathing ambient air or receiving low-flow supplemental oxygen [8]. Further, 5 of 7 patients (71%) who were receiving noninvasive oxygen support (NIPPV or highflow supplemental oxygen) also showed improvement. Only 8 of 53 patients (15%) in the oxygen support category showed worsening health conditions (Figure 3) [8].
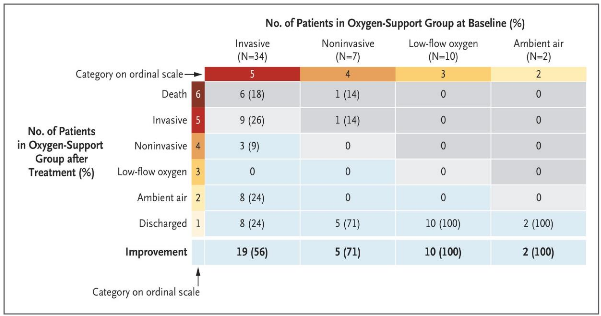
Figure 3: Improvement levels after treatment with remdesivir in patients in various oxygen support category (improvement: blue cells), no change: beige and worsening: gray) [Source: The New England Journal of Medicine (2020)].
Researchers noted that 17 of 30 patients (57%) who were receiving invasive mechanical ventilation were extubated, while 3 of 4 patients (75%) receiving extracorporeal membrane oxygenation (ECMO), or both ECMO and mechanical ventilation stopped receiving it [8]. While the researchers concluded that further tests were needed, these clinical tests showed encouraging results of the therapeutic power of remdesivir to mitigate COVID-19 [8].
An anti-malaria drug, hydroxychloroquine in combination with azithromycin has been shown very promising for repurposing with great potential to treat COVID-19 [9]. It has been known that chloroquine can inhibit pH-dependent steps of the replication of several viruses that can impart a potent effect on SARS-CoV that can help mitigate the infection and spread [1, 7, 10]. Chloroquine is also known to exhibit immunomodulatory effects that help suppress the production/release of TNF-α and IL-6. In addition, researchers have shown that chloroquine can work as a novel class of autophagy inhibitor. The advantage of such an inhibitor is that it may interfere with viral infection and replication. In previous studies, researchers observed the interference of chloroquine with the glycosylation of cellular receptors of SARSCoV that functioned at both entry and at postentry stages of the COVID-19 infection in Vero E6 cells [1, 10, 11]. Chloroquine affects the glycosylation process of angiotensinconverting enzyme 2, known as ACE-2, receptor for binding of viral spike protein, which is essential for interaction with the host [7, 10]. Thus, it is believed that the proven anti-viral and anti-inflammatory activities of chloroquine can be leveraged for its potent efficacy in treating patients with COVID-19 [7]. For example, in a recent clinical report, chloroquine phosphate was recommended to treat COVID-19 associated pneumonia in larger populations [11].
Innovations in the combination antiviral drugs seem to hold the key. To this end, a recent work showed the feasibility of the combination of remdesivir and chloroquine that was demonstrated to effectively inhibit the SARS-CoV-2 in-vitro (Figure 4) [10]. Additionally, it was shown that treatment with chloroquine prevents the spread of SARSCoV infection in the the post-infection period [10]. In this study, researchers employed clinical isolates of SARS-CoV-2, and evaluated the efficacy of seven agents (ribavirin, penciclovir, nitazoxanide, nafamostat, chloroquine, remdesivir [GS5734], and favipiravir (T-705)) in in-vitro conditions (Figure 4) [10]. Cytotoxicity was also evaluated in vero E6 cells, which was followed by infection of the cells with SARSCoV-2 clinical isolates. The test drug was then evaluated at different doses [10]. Reverse transcription PCR-based quantification was done to get the viral yield, which was later confirmed by immunofluorescence microscopy (nucleocapsid protein visualization) (Figure 4) [10]. The results showed that both chloroquine and remdesivir inhibited virus infection at micromolar level (0.77–1.13 μM) and with high selectivity [10].

Figure 4: (a) The in-vitro antiviral activities of the test drugs against SARS-CoV-2 with a Vero E6 cells that were infected with SARS-CoV-2. Viral yield in the cell supernatant was then quantified by qRT-PCR. (b) Immunofluorescence microscopy of virus infection upon treatment of remdesivir and chloroquine. (c & d) Time-of-addition of remdesivir and chloroquine showing NP expression in infected cells that was analyzed by Western blot [Source: Cell Res (2020)]
Concluding Remarks
With the rapid spread of COVID-19 pandemic across the globe, it is of immense importance and absolutely essential to focus on the innovations in discovering drugs to mitigate the challenge. This should be done in addition to the ongoing efforts to develop vaccines for COVID-19 and early diagnostics for rapid testing capabilities and having a robust medical systems including sufficient personal protective equipment for front line healthcare professionals. This should help in preparing to combat the future epidemic outbreaks.
References for further reading
1. Guo, Y., Cao, Q., Hong, Z. et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status, Military Med Res 7, 11 (2020), DOI:10.1186/s40779-020-00240-0
2. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, (2020), DOI https://doi.org/10.1038/s41586-020- 2012-7
3. Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill., 25(4):2000058 (2020), https://doi.org/10.2807/1560- 7917.ES.2020.25.4.2000058
4. Liu Y, Gayle AA, Wilder-Smith A, Rocklov J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med., (2020), https://doi.org/10.1093/jtm/taaa021
5. Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY, Coronaviruses – drug discovery and therapeutic options, Nat Rev Drug Discov, 15 (5): 327–47 (2016), https://doi.org/10.1038/nrd.2015.37
6. Guangdi Li & Erik De Clercq, Therapeutic options for the 2019 novel coronavirus (2019-nCoV), Nat Rev Drug Discov, 19, 149-150 (2020), DOI:10.1038/d41573- 020-00016-0
7. Suliman Khan, Rabeea Siddique, Muhammad Adnan Shereen, Ashaq Ali, Jianbo Liu, Qian Bai, Nadia Bashir, Mengzhou Xue, The emergence of a novel coronavirus (SARS -CoV – 2 ), their biology and therapeutic 2 options, J. Clin. Microbiol. (2020), https://doi.org/10.1128/JCM.00187-20
8. J. Grein, N. Ohmagari, D. Shin, G. Diaz, E. Asperges, A. Castagna, T. Feldt, G. Green et. al., Compassionate Use of Remdesivir for Patients with Severe Covid-19, The New England Journal of Medicine, April 10 (2020), DOI: 10.1056/NEJMoa2007016
9. Philippe GautretJean-Christophe Lagier Philippe Parola Van Thuan Hoang Line Meddeb et. al., Hydroxychloroquine and azithromycin as a treatment of COVID19: results of anopen-label nonrandomized clinical trial, International Journal of Antimicrobial Agents (2020), https://doi.org/10.1016/j.ijantimicag.2020. 105949
10. Wang, M., Cao, R., Zhang, L. et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res., 30, 269–271 (2020), https://doi.org/10.1038/s41422-020- 0282-0
11. Gao J, Tian Z, Yang X, Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID19 associated pneumonia in clinical studies, Biosci Trends. Mar 16;14(1):72- 73 (2020), DOI:10.5582/bst.2020.01047